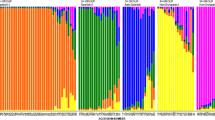
Abstract
Cucurbita pepo is a worldwide cultivated vegetable of American origin. Most of the widely grown commercial types are known as summer squashes and belong to the elongated forms of C. pepo ssp. pepo (Cocozelle, Vegetable marrow and Zucchini groups). These forms were developed in Europe after the arrival of the first American landraces through a process of selection and fixation that led to a loss of genetic diversity. Part of the genetic variability of the first American cultigens remains intact in diverse landraces that are still cultivated for self-consumption and sale in local markets. Using the first collection of genomic and EST-derived microsatellites that has just become available for the species, we compared the natural variation present in a collection of Spanish landraces with that of a set of commercial varieties and hybrids, representing current summer squash market offerings. A total of 194 alleles allowed us to distinguish all the genotypes, even those that were closely related. In general, Cocozelle and Vegetable marrow, groups with considerably long histories, were more variable than the Zucchini group, of more recent origin. We found significant genetic diversity among landraces. The variation present among landraces belonging to the Zucchini group was larger than that of the commercial cultivars. Cluster, principal coordinate and population structure results suggested that the variation of the Spanish landraces has not been extensively used in breeding. Commercial summer squashes can therefore benefit from this underexploited variability, especially from certain landraces that already display favourable commercial traits.



Similar content being viewed by others
References
Blanca J, Cañizares J, Roig C, Ziarsolo P, Nuez F, Picó B (2011) Transcriptome characterization and high throughput SSRs and SNPs discovery in Cucurbita pepo (Cucurbitaceae). BMC Genomics 10(12):104
Boualem A, Fergany M, Fernandez R, Troadec C, Martin A, Morin H, Sari MA, Collin F, Flowers JM, Pitrat M, Purugganan MD, Dogimont C, Bendahmane A (2008) A conserved mutation in an ethylene biosynthesis enzyme leads to andromonoecy in melons. Science 321:836–838
Decker DS (1988) Origin(s), evolution, and systematics of Cucurbita pepo (Cucurbitaceae). Econ Bot 42:4–15
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Esteras C, Nuez F, Picó B (2011) Genetic diversity studies in Cucurbits using molecular tools. In: Wang Y, Behera TK (eds) Cucurbits: genetics, genomics and breeding in crop plants. Science Publishers Inc., Enfield, p 25
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Ezura H, Fukino N (2009) Research tools for functional genomics in melon (Cucumis melo L.): current status and prospects. Plant Biotechnol 26:359–368
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using mulitlocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Falush D, Stephens M, Pritchard JK (2007) Inference of population structure using mulitlocus genotype data: dominant markers and null alleles. Mol Ecol Notes 7:574–578
Fergany M, Balvir Kaur, Monforte AJ, Pitrat M, Rys C, Lecoq H, Dhillon NPS, Dhaliwal SS (2011) Variation in melon (Cucumis melo) landraces adapted to the humid tropics of southern India. Genet Resour Crop Evol 58:225–243
Fernandez-Silva I, Eduardo I, Blanca J, Esteras C, Pico B, Nuez F, Arus P, Garcia-Mas J, Monforte AJ (2008) Bin mapping of genomic and EST-derived SSRs in melon (Cucumis melo L.). Theor Appl Genet 118:139–150
Ferriol M, Pico B (2008) Pumpkin and Winter Squash. In: Prohens J, Nuez F (eds) Handbook of plant breeding, vegetables I. Springer, New York, pp 317–349
Ferriol M, Pico B, Nuez F (2003) Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor Appl Genet 107:271–282
Formisano G, Paris HS, Frusciante L, Ercolano MR (2010) Commercial Cucurbita pepo squash hybrids carrying disease resistance introgressed from Cucurbita moschata have high genetic similarity. Plant Genet Res 8:198–203
Gong L, Stift G, Kofler R, Pachner M, Lelley T (2008) Microsatellites for the genus Cucurbita and an SSR-based genetic linkage map of Cucurbita pepo L. Theor Appl Genet 117:37–48
Gonzalez VM, Rodríguez-Moreno L, Centeno E, Benjak A, Garcia-Mas J, Puigdomènech P, Aranda MA (2010) Genome-wide BAC-end sequencing of Cucumis melo using two BAC libraries. BMC Genomics 11:618
Huang S, Li R, Zhang Z, Li L, Gu X, Fan W, Lucas WJ, Wang X, Xie B, Ni P, Ren Y, Zhu H, Li J, Lin K, Jin W, Fei Z, Li G, Staub J, Kilian A, van der Vossen EA, Wu Y, Guo J, He J, Jia Z, Ren Y, Tian G, Lu Y, Ruan J, Qian W, Wang M, Huang Q, Li B, Xuan Z, Cao J, Asan, Wu Z, Zhang J, Cai Q, Bai Y, Zhao B, Han Y, Li Y, Li X, Wang S, Shi Q, Liu S, Cho WK, Kim JY, Xu Y, Heller-Uszynska K, Miao H, Cheng Z, Zhang S, Wu J, Yang Y, Kang H, Li M, Liang H, Ren X, Shi Z, Wen M, Jian M, Yang H, Zhang G, Yang Z, Chen R, Liu S, Li J, Ma L, Liu H, Zhou Y, Zhao J, Fang X, Li G, Fang L, Li Y, Liu D, Zheng H, Zhang Y, Qin N, Li Z, Yang G, Yang S, Bolund L, Kristiansen K, Zheng H, Li S, Zhang X, Yang H, Wang J, Sun R, Zhang B, Jiang S, Wang J, Du Y, Li S (2009) The genome of the cucumber, Cucumis sativus L. Nat Genet 41:1275–1281
Hubisz MJ, Falush D, Stephens M, Pritcard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Resour 9:1322–1332
Lebeda A, Widrlechner MP, Staub J, Ezura H, Zalapa J, Kristkova E (2007) Cucurbits (Cucurbitaceae; Cucumis spp., Cucurbita spp., Citrullus spp.). In: Singh RJ (ed) Genetic resources, chromosome engineering, and crop improvement, vol 3. CRC Press, Boca Raton, pp 271–376
Li Z, Huang S, Liu S, Pan J, Zhang Z, Tao Q, Shi Q, Jia Z, Zhang W, Chen H, Si L, Zhu L, Cai R (2009) Molecular isolation of the M gene suggests that a conserved-residue conversion induces the formation of bisexual flowers in cucumber plants. Genetics 182:1381–1385
Lira R, Montes S (1994) Cucurbits (Cucurbita spp.). In: Hernandez JE, Leon J (eds) Neglected crops, 1492 from a different perspective. F.A.O, Rome, pp 63–77
Liu K, Muse SV (2005) PowerMarker: integrated analysis environment for genetic marker analysis. Bioinformatics 21:2128–2129
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Metzker ML (2010) Applications of next-generation sequencing. Sequencing technologies the next generation. Nat Rev Genet 11:31–46
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Nesom GL (2011) Toward consistency of taxonomic rank in wild/domesticated Cucurbitaceae. Phytoneuron 13:1–33
Paris HS (1986) A proposed subspecific classification for Cucurbita pepo. Phytologia 61:133–138
Paris HS (2000) History of the cultivar-groups of Cucurbita pepo. In: Janick J (ed) Hort revs, vol 25. Wiley, New York, pp 71–170
Paris HS (2008) Summer squash. In: Prohens J, Nuez F (eds) Handbook of plant breeding, vegetables I. Springer, New York, pp 351–379
Paris HS, Brown RN (2005) The genes of pumpkin and squash. HortScience 40:1620–1630
Paris HS, Yonash N, Portnoy V, Mozes-Daube N, Tzuri G, Katzir N (2003) Assessment of genetic relationships in 4 Cucurbita pepo (Cucurbitaceae) using DNA markers. Theor Appl Genet 106:971–978
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rohlf JF (1998) NTSYS: numerical taxonomy and multivariate analysis system, version 2.02. Exeter Software, Setauket
Sanjur OI, Piperno DR, Andres TC, Wessel-Beaver L (2002) Phylogenetic relationships among domesticated and wild species of Cucurbita (Cucurbitaceae) inferred from a mitochondrial gene: implications for crop plant evolution and areas of origin. Proc Natl Acad Sci USA 99:535–540
Smith BD (1997) The initial domestication of Cucurbita pepo in the Americas 10,000 years ago. Science 276:932–934
Acknowledgments
This research was funded by Projects INIA (RTA2008-00035-C02-02 and RTA2011-00044-C02-2) of the Spanish Instituto Nacional de Investigación y Tecnología Agraria. G. Formisano was supported by the Italian Doctorate School in Agrobiology and Agrochemistry of the University of Naples “Federico II”. Authors want to thank E. Martínez Pérez her technical assistance for fruit characterization.
Author information
Authors and Affiliations
Corresponding author
Additional information
Gelsomina Formisano and Cristina Roig have contributed equally to this work.
Rights and permissions
About this article
Cite this article
Formisano, G., Roig, C., Esteras, C. et al. Genetic diversity of Spanish Cucurbita pepo landraces: an unexploited resource for summer squash breeding. Genet Resour Crop Evol 59, 1169–1184 (2012). https://doi.org/10.1007/s10722-011-9753-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-011-9753-y