Abstract
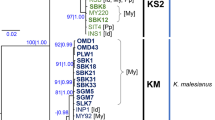
Unraveling the cryptic genetic diversity and selective breeding network in various Porphyra strains is of significance for conservation and utilization of economically important nori crops, for both current and future needs. Here, we used nuclear ribosomal spacer (ITS1) region to investigate the genetic variation and intra-specific relatedness of 59 Porphyra yezoensis Ueda specimens worldwide using phylogenetic and parsimony genealogical approaches. 23 nrDNA ITS1 genotypes were revealed and clustering analysis grouped them into two distinct clades. High genetic diversity was detected in wild P. yezoensis strains from Miyagi and Hokkaido Prefectures in Japan, while the cultivated strains from China and South Korea exhibited relatively higher genetic diversity likewise, which provided crucial genetic insights for future commercial breeding of P. yezoensis on a global scale. In addition, phylogenetic study has revealed the genetic relationship of strains with unknown parentage to those with known parentage, and also ITS1 sequence pattern could be correlated with the geographic origin of P. yezoensis specimens. All these pedigree information generated from this research can be used to select parents for inter-specific or intra-specific selective breeding and cross breeding to maximize the preservation of stock resource and sustainable development of nori industry.


Similar content being viewed by others
References
Acinas SG, Sarma-Rupavtarm R, Klepac-Ceraj V, Polz MF (2005) PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed the same sample. Appl Environ Microbiol 71:8966–8969
Ainouche ML, Bayes RJ (1997) On the origins of the tetraploid Bromus species (section Bromus, Poaceae): insights from the internal transcribed spacer sequences of nuclear ribosomal DNA. Genome 40:730–743
Baldwin BG, Sanderson MJ, Porter JM, Wojciechowski MF, Campbell CS, Donoghue MJ (1995) The ITS region of nuclear ribosomal DNA: a valuable source of evidence on angiosperm phylogeny. Ann Mo Bot Gard 82:247–277
Clement M, Posada D, Crandall K (2000) TCS: a computer program to estimate gene genealogies. Mol Ecol 9:1657–1659
Coyer JA, Peters AF, Hoarau G, Stam WT, Olesn JL (2002) Hybridization of the marine seaweeds, Fucus serratus and F. evanescens (Heterokontophyta: Phaeophyceae) in a century-old zone of secondary contact. Proc R Soc Lond Biol Sci 269:1829–1834
Coyer JA, Hoarau G, Stam WT, Olsen JL (2007) Hybridization and introgression in a mixed population of the intertidal seaweeds Fucus evanescens and F. serratus. J Evol Biol 20:2322–2333
Feliner GN, Larena BG, Aguilar JF (2004) Fine scale geographic structure, intra-individual polymorphism and recombination in nuclear ribosomal internal transcribed spacers in Armeria (Plumbaginaceae). Ann Bot 93:189–200
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–793
Hu ZM, He YJ, Xia P, Duan DL (2007) Molecular identification of Chinese cultivated Porphyra (Bangiaceae, Rhodophyta) based on the rDNA internal transcribed spacer-1 sequence and random amplified polymorphic DNA markers. Mar Biol Res 3:20–28
Huelsenbeck JP, Crandall KA (1997) Phylogeny estimation and hypothesis testing using maximum likelihood. Ann Rev Ecol Syst 28:437–466
Hwang MS, Kim SM, Ha DS, Baek JM, Kim HS, Choi HG (2005) DNA sequences and identification of Porphyra cultivated by natural seeding on the Southwest coast of Korea. Algae 20:183–196 (in Korean with English abstract)
Iitsuka O, Nakamura K, Ozaki A, Okamoto N, Saga N (2002) Genetic information of three pure lines of Porphyra yezoensis (Bangiales, Rhodophyta) obtained by AFLP analysis. Fish Sci 68:1113–1117
Keirstein G, Vallinoto M, Silva A, Schneider MP, Iannuzzi L, Brenig B (2004) Analysis of mitochondrial D-loop region casts new light on domestic water buffalo (Bubalus bubalis) phylogeny. Mol Phylogenet Evol 30:308–324
Kunimoto M, Kito H, Kaminishi Y, Mizukami Y, Murase N (1999) Molecular divergence of the SSU rRNA gene and internal transcribed spacer 1 in Porphyra yezoensis (Rhodophyta). J Appl Phycol 11:211–216
Kunimoto M, Kito H, Mizukami Y, Murase N, Levine I (2003) Molecular features of a defined genetic marker for the determination of the Porphyra tenera lineage. J Appl Phycol 15:337–343
Miura A (1988) Taxonomic studies of Porphyra species cultivated in Japan, referring to their transition to the cultivated variety. J Tokyo Univ Fish 75:311–325
Miura A (1998) Asakusa-nori: data book of rare aquatic animals and plants of Japan, edited by Japan Fisheries Resource Conservation Association. Japan Fisheries Resource Conservation Association, Tokyo, pp 298–299 (in Japanese)
Miura A, Aruga Y (1987) Distribution of Porphyra in Japan as affected by cultivation. J Tokyo Univ Fish 74:41–50
Mizukami Y, Kito H, Kaminishi Y, Murase N, Kunimoto M (1999) Nucleotide sequence variation in the ribosomal internal transcribed spacer regions of cultivated (cultivars) and field-collected thalli of Porphyra yezoensis. Fish Sci 65:788–789
Neefus CD, Mathieson A, Bray TL, Yarish C (2008) The distribution, morphology, and ecology of three introduced Asiatic species of Porphyra (Bangiales, Rhodophyta) in the Northwestern Atlantic. J Phycol 44:1399–1414
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Nei M, Li WH (1979) Mathematical model for studying genetic variation in terms of restriction endonucleases. Proc Natl Acad Sci USA 76:5269–5273
Niwa K, Aruga Y (2003) Rapid DNA extraction from conchocelis and ITS-1 rDNA sequences of seven strains of cultivated Porphyra yezoensis (Bangiales, Rhodophyta). J Appl Phycol 15:29–33
Niwa K, Aruga Y (2006) Identification of currently cultivated Porphyra species by PCR-RFLP analysis. Fish Sci 72:143–148
Niwa K, Kikuchi N, Iwabuchi M, Aruga Y (2004) Morphological and AFLP variation of Porphyra yezoensis Ueda form narawaensis Miura (Bangiales, Rhodophyta). Phycol Res 52:180–190
Niwa K, Kato A, Kobiyama A, Kawai H, Aruga Y (2008) Comparative study of wild and cultivated Porphyra yezoensis (Bangiales, Rhodophyta) based on molecular and morphological data. J Appl Phycol 20:261–270
Niwa K, Iida S, Kato A, Kawai H, Kikuchi N, Kobiyama A, Aruga Y (2009) Genetic diversity and introgression in two cultivated species (Porphyra yezoensis and Porphyra tenera) and closely related wild species of Porphyra (Bangiales, Rhodophyta). J Phycol 45:493–502
Posada D, Crandall KA (1998) Modeltest: testing the model of DNA substitution. Bioinformatics 14:817–818
Posada D, Crandall KA (2001) Intraspecific gene genealogies: trees grafting into networks. Trends Ecol Evol 16:37–45
Rozas J, Rozas R (1999) DnaSP version 3: an integrated program for molecular population genetics and molecular evolution analysis. Bioinformatics 15:174–175
Swofford DL (2002) PAUP: phylogenetic analysis using parsimony (and other methods). Version 4.0b10. Sinauer, Sunderland
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Wallace A, Klein AS, Mathieson AC (2004) Determing the affinities of salt march fucoids using microsatellite markers: evidence of hybridization and introgression between two species of Fucus (Phaeophyta) in a Maine estuary. J Phycol 40:1013–1027
Watterson GA (1975) On the number of segregating sites in genetical models without recombination. Theor Pop Biol 7:256–276
Weng ML, Liu B, Jin DM, Yang QK, Zhao G, Ma JH, Xu P, Duan DL, Wang B (2005) Identification of 27 Porphyra lines (Rhodophyta) by DNA fingerprinting and molecular markers. J Appl Phycol 17:91–97
Won H, Renner SS (2005) The internal transcribed spacer of nuclear ribosomal DNA in the gymnosperm Gnetum. Mol Phylogenet Evol 36:581–597
Yoshida T (1998) Marine algae of Japan. Uchida Rokakuho, Tokyo, p 222 (in Japanese)
Yoshida T (2000) Porphyra tenera Kjellman: threatened wildlife of Japan, edited by Environment Agency of Japan, 2nd edn. Japan Wildlife Research Center, Tokyo, p 218 (in Japanese)
Acknowledgments
The authors thank anonymous reviewers for insightful criticisms and comments on an earlier version of this manuscript. This research was partly supported by China Postdoctoral Science Foundation project (No. 20090451356) and Knowledge Innovation Program of the Chinese Academy of Sciences on Biodiversity Project.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Hu, Z., Liu, F., Shao, Z. et al. NrDNA internal transcribed spacer revealed molecular diversity in strains of red seaweed Porphyra yezoensis and genetic insights for commercial breeding. Genet Resour Crop Evol 57, 791–799 (2010). https://doi.org/10.1007/s10722-009-9519-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-009-9519-y