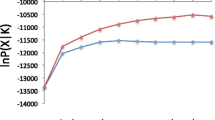
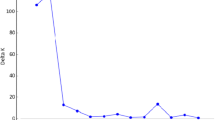
Abstract
Iwateyamanashi (Pyrus ussuriensis var. aromatica) is one of the Pyrus species which grows wild in Japan. The number of Iwateyamanashi trees has been decreasing, so conservation and evaluation is urgently needed. Over 500 accessions of Pyrus species collected from Iwate in northern Tohoku region are maintained at Kobe University as an Iwateyamanashi germplasm collection. In order to investigate the genetic diversity, five SSR (simple sequence repeat) markers, developed from Japanese and European pear were examined for 86 Pyrus individuals including 58 accessions from Iwate. These SSR loci could discriminate between all the Iwate accessions except for 10 that bear seedless fruit, as well as determine the genetic diversity in Iwateyamanashi germplasms. High levels of variation were detected in 41 alleles and the mean observed heterozygosity across 5 loci was 0.50 for the Iwate accessions. Seedless accessions sharing identical SSR genotype with the local pear variety “Iwatetanenashi” were supposed to have been propagated vegetatively via grafting. In an UPGMA phenogram, Japanese pear varieties (P. pyrifolia) were clustered into two groups with some Iwate accessions including seedless ones. Another 38 Iwate accessions were not clustered clearly, and there was no clear relationship between these accessions and geographical distribution or morphological characters. Allele frequency revealed that the Iwate accessions were genetically more divergent than the Japanese pear varieties. Most Japanese pears possessed a 219 bp deletion at a spacer region between the accD and psaI genes in the chloroplast DNA (cpDNA), but other Pyrus species and two Iwateyamanashi trees did not. In the Iwate accessions, 79.3% had a deletion type cpDNA and others had a standard type cpDNA without deletion. These results are indicative of the wide range of genetic diversity in the Iwate accessions which include Japanese pear varieties. A combination of SSR and cpDNA analyses revealed high heterogeneity in Iwateyamanashi and coexistence of Iwateyamanashi and hybrid progeny with P. pyrifolia. These could be reasons for the wide range of continuous morphological variation described previously.




Similar content being viewed by others
Abbreviations
- FRC-KU:
-
Food Resources Education and Research Center, Kobe University
- BG-OCU:
-
Botanical Gardens, Osaka City University
- NIFTS:
-
National Institute of Fruit Tree Science
References
Asami Y (1921) Nipponnashi no shurui. Nippon Engei Zasshi 33:1–11
Bell RL, Hough LF (1986) Interspecific and intergeneric hybridization of Pyrus. HortScience 21:62–64
Brookfield JF (1996) A simple new method for estimating null allele frequency from heterozygote deficiency. Mol Ecol 5:453–455
Challice JS, Westwood MN (1973) Numerical taxonomic studies of the genus Pyrus using both chemical and botanical characters. Bot J Linn Soc 67:121–148
Hokanson SC, Szewc-McFadden AK, Lamboy WF, McFerson JR (1998) Microsatellite (SSR) markers reveal genetic identities, genetic diversity and relationships in a Malus × domestica Borkh. core subset collection. Theor Appl Genet 97:671–683
Hokanson SC, Lamboy WF, Szewc-McFadden AK, McFerson JR (2001) Microsatellite (SSR) variation in a collection of Malus (apple) species and hybrids Euphytica 118:281–294
Hosaka K (1995) Successive domestication and evolution of the Andean potatoes as revealed by chloroplast DNA restriction endnuclease analysis. Theor Appl Genet 90:356–363
Iketani H, Manabe T, Matsuta N, Akihama T, Hayashi T (1998) Incongruence between RFLPs of chloroplast DNA and morphological classification in east Asia pear (Pyrus spp.). Genet Resour Crop Evol 45:533–539
Iketani H, Ohashi H (2003) Taxonomy and distribution of Japanese populations of Pyrus ussuriensis Maxim. (Rosaceae). J Jpn Bot 78:119–134
Kajiura I (1983) Nihon nashi no kigen to chiriteki bunka. In: Akihama T (ed) Ikushugaku Saikin no Shinpo, vol. 25. Youkendo, Tokyo, pp 3–13 (in Japanese)
Kajiura I, Nakajima M, Sakai Y, Oogaki C (1983) A species-specific flavonoid from Pyrus ussuriensis Max. and Pyrus aromatica Nakai et Kikuchi, and its geographical distribution in Japan. Jpn J Breed 33:1–14
Kajiura I, Sato Y (1990) Recent progress in Japanese pear (Pyrus pyrifolia Nakai) breeding, and descriptions of cultivars based on literature review. Bull Fruit Tree Res Sta. Extra No. 1:1–329 (in Japanese)
Katayama H, Uematsu C (2002) Phylogenetic analysis of Pyrus species: the chloroplast DNA structure of Pyrus ussuriensis var. hondoensis. Acta Hortic 587:259–268
Katayama H, Uematsu C (2003) Comparative analysis of chloroplast DNA in Pyrus species: physical map and gene localization. Theor Appl Genet 106:303–310
Katayama H, Uematsu C (2006) Pear (Pyrus species) genetic resources in Iwate, Japan. Genet Resour Crop Evol 53:483–498
Kikuchi A (1948) Nashi. Kajuengeigaku Jyo-kan, Kajyu-syurui kakuron, Yokendo, Tokyo, pp 64–116 (in Japanese)
Kimura T, Shi YZ, Shoda M, Kotobuki K, Matsuta N, Hayashi T, Ban Y, Yamamoto T (2002) Identification of Asian pear varieties by SSR analysis. Breed Sci 52:115–121
Kimura T, Iketani H, Kotobuki K, Matsuta N, Ban Y, Hayashi T, Yamamoto T (2003) Genetic characterization of pear varieties revealed by chloroplast DNA sequences. J Hort Sci & Biot 78:241–247
Koidzumi G (1924) Contributiones ad cognitionem florae Asiae Orientalis. Bot Mag (Tokyo) 38:87–114
Kloosterman AD, Budowle B, Daselaar P (1993) PCR-amplification and detection of the human D1S80 VNTR locus. Amplification conditions, population genetics and application in forensic analysis. Int J Leg Med 105:257–264
Makino T (1908) Observations on the Flora of Japan. Bot Mag (Tokyo) 22:63–72
Matsuyama A (1930) Notes on Pyrus hondoensis, Nakai et Kikuchi. Bull Alumn: Assoc Chiba Hortic Coll 1:17–27 (in Japanese)
Nakai T (1918) Notulae ad plantas Japoniae et Koreae XVI. Bot Mag (Tokyo) 32:28–37
Nakai T (1919) Notulae ad Plantas Japoniae et Koreae XXI Bot Mag (Tokyo) 33:193–216
Nei M (1973) Analysis of genetic diversity in subdivided populations. Proc Natl Acad Sci USA 70:3321–3323
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Ozawa T, Sakuta H, Negishi O, Kajiura I (1995) Identification of species-specific flavone glucosides useful as chemotaxonomic markers in the genus Pyrus. Biosci Biotech Biochem 59:2244–2246
Ohwi J (1965) Flora of Japan, Smithonian Institution, Washington DC, pp 549
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingley S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for germplasm analysis. Mol Breed 2:225–238
Rehder A (1940) Manual of cultivated trees and shrubs, 2nd edn. Macmillan, New York
Rohlf FJ (1998) NTSYS, numerical taxonomy and multivariate analysis system, Ver. 2.01. Exeter Publishing, Ltd., Setauket, New York
Small RL, Ryburn JA, Cronn RC, Seelanan T, Wendel JF (1998) The tortoise and the hare: choosing between noncoding plastome and nuclear ADH sequences for phylogeny reconstruction in recently diverged plant group. Am J Bot 85:1301–1315
Tessier C, David J, This P, Boursiquot JM, Charrier A (1999) Optimization of the choice of molecular markers for variental identification in Vitis vinifera L. Theor Appl Genet 98:171–177
Yamamoto T, Kimura T, Sawamura Y, Kotobuki K, Ban Y, Hayashi T, Matsuta N (2001) SSRs isolated from apple can identify polymorphism and genetic diversity in pear. Theor Appl Genet 102:865–870
Yamamoto T, Kimura T, Sawamura Y, Manabe T, Kotobuki K, Hayashi T, Ban Y, Matsuta N (2002a) Simple sequence repeats for genetic analysis in pear. Euphytica 124:129–137
Yamamoto T, Kimura T, Ban Y, Shoda M, Hayashi T, Matsuta N (2002b) Development of microsatellite markers in Japanese pear (Pyrus pyrifolia Nakai). Mol Ecol Notes 2:14–16
Yamamoto T, Kimura T, Saito T, Kotobuki K, Matsuta N, Liebhard R, Gessler C, Van De Weg WE, Hayashi T (2004) Genetic linkage maps of Japanese and European pears aligned to the apple consensus map. Acta Hortic 663:51–56
Yoneyama K, Makita M (2002) A search for genetic resources on the genes of Pyrus in Japan Iwateyamanashi (P. aromatica Nakai et Kikuchi). Acta Hortic 587:147–150
Acknowledgements
Special thanks to Dr. H. Iketani, National Institute of Fruit Tree Science, and Dr. Anne Edwards, John Innes Centre, UK for their critical readings. Also we thank Dr. K. Hosaka for his suggestions and many technicalities. Thanks to Mr. M. Miyake, Mr. S. Kakehi, Mr. H. Hashizume, Mr. M. Fujimatsu, Mr. S. Kitanishi and Mr. K. Masaki, Kobe University for their plant maintenance. This work was supported in part by a General Research Grant from the Nippon Life Insurance Foundation to C. U. Research Grant from Sanriku foundation, and Grant-in-Aid (No.17510196) for Scientific Research from the Ministry of Education, Science and Culture, Japan.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Katayama, H., Adachi, S., Yamamoto, T. et al. A wide range of genetic diversity in pear (Pyrus ussuriensis var. aromatica) genetic resources from Iwate, Japan revealed by SSR and chloroplast DNA markers. Genet Resour Crop Evol 54, 1573–1585 (2007). https://doi.org/10.1007/s10722-006-9170-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10722-006-9170-9