Abstract
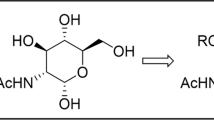
As part of our ongoing program aimed at understanding the structural significance of GlcNAcβAsn linkage conserved in all eukaryotic N-glycoproteins, the present study reports on the synthesis and X-ray crystal structures of N-(3-deoxy-3-acetamido-β-D-glycopyranosyl)acetamide (Glc3NAcβNHAc) and the corresponding propionamide (Glc3NAcβNHPr). Comparative analysis of these structures with those of the corresponding GlcNAc (C2 acetamido) compounds showed that the bifurcated anti-parallel pattern involving N–H…O and C–H…O hydrogen bonds, the hallmark feature of the N-glycoprotein models, GlcNAcβNHAc and GlcNAcβAsn, is absent in both the C3 acetamido analogs. The extended (anti) conformation of the amido aglycon moiety as defined by χ 2 seen in the case of C2 acetamido derivative, GlcNAcβNHPr, turns into gauche for the C3 acetamido analog (Glc3NAcβNHPr). This observation is consistent with the earlier work on the critical role of the C2-NHAc group of GlcNAcβAsn in controlling χ 2 at the linkage region of N-glycoproteins.






Similar content being viewed by others
Abbreviations
- Glc3NAcβNHAc:
-
N-(3-deoxy-3-acetamido-β-D-glycopyranosyl)acetamide
- Glc3NAcβNHPr:
-
N-(3-deoxy-3-acetamido-β-D-glycopyranosyl)propionamide
- NMR:
-
Nuclear Magnetic Resonance
- ESI-MS:
-
Electrospray Ionization Mass Spectrometry
- ORTEP:
-
Oak Ridge Thermal Ellipsoid Plot
References
Varki, A.: Biological roles of oligosaccharides: all of the theories are correct. Glycobiology 3, 97–130 (1993)
Spiro, R.G.: Protein glycosylation: nature, distribution, enzymatic formation, and disease implication of glycopeptide bonds. Glycobiology 12, 43R–56R (2002)
Ohanessian, J., Avenel, D., Neuman, A., Gillier-Pandraud, H.: Structure cristalline de la 2- acétamido-1-N-(L-aspart-4-oyl)-2-désoxy-β-D-glucopyranosylamine. Carbohydr. Res. 80, 1–13 (1980)
Sriram, D., Lakshmanan, T., Loganathan, D., Srinivasan, S.: Crystal structure of a hydrated N-glycoprotein linkage region model and its analogue: hydrogen bonding and π-π stacking driven molecular assembly. Carbohydr. Res. 309, 227–236 (1998)
Lakshmanan, T., Sriram, D., Priya, K., Loganathan, D.: On the structural significance of the linkage region constituents of N-glycoproteins: an X-ray crystallographic investigation using models and analogs. Biochem. Biophys. Res. Commun. 312, 405–413 (2003)
Loganathan, D., Aich, U.: Observation of a unique pattern of bifurcated hydrogen bonds in the crystal structures of the N-glycoprotein linkage region models. Glycobiology 16, 343–348 (2006)
Cioci, G., Srivastava, A., Loganathan, D., Mason, S.A., Pérez, S., Imberty, A.: Low temperature neutron diffraction structures of N-glycoprotein linkage models and analogs: structure refinement and trifurcated hydrogen bonds. J. Am. Chem. Soc. 133, 10042–10045 (2011)
Mohamed Naseer Ali, M., Aich, U., Varghese, B., Pérez, S., Imberty, A., Loganathan, D.: Conformational preferences of the aglycon moiety in models and analogs of GlcNAc-Asn linkage: crystal structures and ab initio quantum chemical calculations of N-(β-D -glycopyranosyl)haloacetamides. J. Am. Chem. Soc. 130, 8317–8325 (2008)
Muhizi, T., Grelier, S., Coma, V.: Synthesis and antibacterial activity of aminodeoxyglucose derivatives against Listeria innocua and Salmonella typhimurium. J. Agric. Food Chem. 57, 8770–8775 (2009)
Barton, D.H.R., Jaszberenyi, J.C., Theodorakis, E.A., Reibenspiest, J.H.: The invention of radical reactions. 30. diazirines as carbon radical traps. Mechanistic aspects and synthetic applications of a novel and efficient amination process. J. Am. Chem. Soc. 115, 8050–8059 (1993)
Bruker SADABS, Bruker AXS Inc., Madison, Wisconsin, USA (1999)
Sheldrick, G.M. SHELXL97, Program for Crystal Structure Refinement; University of Gottingen, Germany (1997)
Farrugia, L.J.: ORTEP-3 for Windows-a version of ORTEP-III with a Graphical User Interface (GUI). J. Appl. Cryst. 30, 565 (1997)
Bruno, I.J., Cole, J.C., Edgington, P.R., Kessler, M., Macrae, C.F., McCabe, P., Pearson, J., Taylor, R.: New software for searching the Cambridge Structural Database and visualizing crystal structure. Acta Cryst. B58, 389–397 (2002)
Delbaere, L.T.J.: The molecular and crystal structures of 4-N-(2-acetamido-2-deoxy-β-D -glucopyranosyl)-L-asparagine trihydrate and 4-N-(β-D-glucopyranosyl)-L-asparagine monohydrate. The X-ray analysis of a carbohydrate-peptide linkage. Biochem. J. 143, 197–205 (1974)
Marchessault, R.H., Perez, S.: Conformations of the hydroxymethyl group in crystalline aldohexopyranoses. Biopolymers 18, 2369–2374 (1979)
Larkin, A., Imperiali, B.: The expanding horizons of asparagine-linked glycosylation. Biochemistry 50, 4411–4426 (2011)
Acknowledgment
The funding provided by Department of Science and Technology (DST), New Delhi for the purchase of the 400 MHz NMR under IRHPA Scheme and ESI-MS under the FIST program to the Department of Chemistry, IIT Madras is gratefully acknowledged. The authors thank the single crystal XRD facility, Chemistry Department, IIT Madras for the X-ray data collection. We are thankful to Mr. V. Ram Kumar for X-ray data collection. One of us (M.M) is thankful to the Council of Scientific and Industrial Research (CSIR), New Delhi, for the award of a Senior Research Fellowship. We also thank Cambridge Crystallographic Data Centre (CCDC), United Kingdom, for making the program Mercury 2.3 available for use.
Supplementary
Data Complete structural data of the Glc3NAcβNHAc (3) and Glc3NAcβNHPr (4) have been deposited at the Cambridge Crystallographic Data Centre (CCDC # 840488 – 840489, respectively), and can be obtained free of charge via www.ccdc.cam.ac.uk/data_request/cif (or from the Director, Cambridge Crystallographic Data Centre, 12 Union Road, Cambridge CB2 1EZ, UK; fax: +44-1223-336033; or email: deposit@ccdc.cam.ac.uk).
Author information
Authors and Affiliations
Corresponding author
Additional information
Dedicated to late Prof. Nathan Sharon
Rights and permissions
About this article
Cite this article
Mathiselvam, M., Varghese, B. & Loganathan, D. Synthesis and X-ray crystallographic investigation of N-(3-deoxy-3-acetamido-β-D-glycopyranosyl)alkanamides as analogs of N-glycoprotein linkage region. Glycoconj J 28, 573–580 (2011). https://doi.org/10.1007/s10719-011-9357-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10719-011-9357-y