Abstract
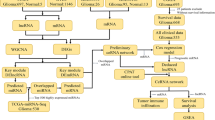
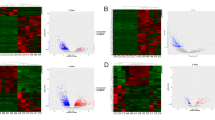
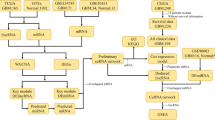
Messenger RNA (mRNA) and long noncoding RNA (lncRNA) targets interact via competitive microRNA (miRNA) binding. However, the roles of cancer-specific lncRNAs in the competing endogenous RNA (ceRNA) networks of low-grade glioma (LGG) remain unclear. This study obtained RNA sequencing data for normal solid tissue and LGG primary tumour tissue from The Cancer Genome Atlas database. We used a computational method to analyse the relationships among the mRNAs, lncRNAs, and miRNAs in these samples. Gene ontology (GO) function and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis was used to predict the biological processes (BPs) and pathways associated with these genes. Kaplan–Meier survival analysis was used to evaluate the association between the expression levels of specific mRNAs, lncRNAs, and miRNAs and overall survival. Finally, we created a ceRNA network describing the relationships among these mRNAs, lncRNAs, and miRNAs using Cytoscape 3.5.1. A total of 2555 differentially expressed (DE) mRNAs, 218 DElncRNAs, and 192 DEmiRNAs were identified using R. In addition, GO and KEGG pathway analysis of the mRNAs and lncRNAs in the ceRNA network identified 10 BPs, 10 cell components, 10 molecular functions, and 48 KEGG pathways as selectively enriched. A total of 55 lncRNAs, 50 miRNAs, and 10 mRNAs from this network were shown to be closely associated with overall survival in LGG. Finally, 59 miRNAs, 235 mRNAs, and 17 lncRNAs were used to develop a ceRNA network comprising 313 nodes and 1046 edges. This study helps expand our understanding of ceRNA networks and serves to clarify the underlying pathogenesis mechanism of LGG.




Similar content being viewed by others
Data availability
We obtained data from TCGA database (https://portal.gdc.cancer.gov/).
References
Buckner JC, Shaw EG, Pugh SL, Chakravarti A, Gilbert MR et al (2016) Radiation plus procarbazine, CCNU, and vincristine in low-grade glioma. N Engl J Med 374:1344–1355
Chen L, Wang X, Wang H, Li Y, Yan W et al (2012) miR-137 is frequently down-regulated in glioblastoma and is a negative regulator of Cox-2. Eur J Cancer 48:3104–3111
Chou CH, Chang NW, Shrestha S, Hsu SD, Lin YL et al (2016) miRTarBase 2016: updates to the experimentally validated miRNA-target interactions database. Nucleic Acids Res 44:D239-247
Deng D, Xue L, Shao N, Qu H, Wang Q et al (2016) miR-137 acts as a tumor suppressor in astrocytoma by targeting RASGRF1. Tumour Biol 37:3331–3340
Fan Q, Liu B (2018) Comprehensive analysis of a long noncoding RNA-associated competing endogenous RNA network in colorectal cancer. Onco Targets Ther 11:2453–2466
Fang XN, Yin M, Li H, Liang C, Xu C et al (2018) Comprehensive analysis of competitive endogenous RNAs network associated with head and neck squamous cell carcinoma. Sci Rep 8:10544
Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE et al (2015) A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 49:213–242
Fu Q, Li S, Zhou Q, Yalikun K, Yisireyili D et al. (2019) Low LINC00599 expression is a poor prognostic factor in glioma. Biosci Rep 39
Gao Y, Li L, Zheng H, Zhou C, Chen X et al (2020) KIF3C is associated with favorable prognosis in glioma patients and may be regulated by PI3K/AKT/mTOR pathway. J Neurooncol 146:513–521
Guo JU, Agarwal V, Guo H, Bartel DP (2014) Expanded identification and characterization of mammalian circular RNAs. Genome Biol 15:409
Hsu JB, Chang TH, Lee GA, Lee TY, Chen CY (2019) Identification of potential biomarkers related to glioma survival by gene expression profile analysis. BMC Med Genom 11:34
Huang J, Jiang W, Tong X, Zhang L, Zhang Y et al (2019) Identification of gene and microRNA changes in response to smoking in human airway epithelium by bioinformatics analyses. Medicine 98:e17267
Huberfeld G, Vecht CJ (2016) Seizures and gliomas–towards a single therapeutic approach. Nat Rev Neurol 12:204–216
Kwok ZH, Tay Y (2017) Long noncoding RNAs: lincs between human health and disease. Biochem Soc Trans 45:805–812
Li CY, Liang GY, Yao WZ, Sui J, Shen X et al (2016) Integrated analysis of long non-coding RNA competing interactions reveals the potential role in progression of human gastric cancer. Int J Oncol 48:1965–1976
Li JH, Liu S, Zhou H, Qu LH, Yang JH (2014) starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res 42:D92-97
Li KK, Yang L, Pang JC, Chan AK, Zhou L et al (2013) MIR-137 suppresses growth and invasion, is downregulated in oligodendroglial tumors and targets CSE1L. Brain Pathol 23:426–439
Liang W, Sun F (2019) Competing endogenous RNA network analysis reveals pivotal ceRNAs in adrenocortical carcinoma. Front Endocrinol 10:301
Lin P, Wen DY, Li Q, He Y, Yang H et al (2018) Genome-wide analysis of prognostic lncRNAs, miRNAs, and mRNAs forming a competing endogenous RNA network in hepatocellular carcinoma. Cell Physiol Biochem 48:1953–1967
Ling H, Fabbri M, Calin GA (2013) MicroRNAs and other non-coding RNAs as targets for anticancer drug development. Nat Rev Drug Discov 12:847–865
Liu B, Ma T, Li Q, Wang S, Sun W et al (2019) Identification of a lncRNA-associated competing endogenous RNA-regulated network in clear cell renal cell carcinoma. Mol Med Rep 20:485–494
Long S, Li G (2019) Comprehensive analysis of a long non-coding RNA-mediated competitive endogenous RNA network in glioblastoma multiforme. Exp Ther Med 18:1081–1090
Ma K, Zhao Q, Li S (2014) Competing endogenous RNA network in pulmonary arterial hypertension. Int J Cardiol 172:e527-528
Nibbe RK, Chowdhury SA, Koyutürk M, Ewing R, Chance MR (2011) Protein-protein interaction networks and subnetworks in the biology of disease. Wiley Interdiscip Rev Syst Biol Med 3:357–367
Olar A, Sulman EP (2015) Molecular markers in low-grade glioma-toward tumor reclassification. Semin Radiat Oncol 25:155–163
Pallud J, Audureau E, Blonski M, Sanai N, Bauchet L et al (2014) Epileptic seizures in diffuse low-grade gliomas in adults. Brain 137:449–462
Rieder CR, Parsons RB, Fitch NJ, Williams AC, Ramsden DB (2000) Human brain cytochrome P450 1B1: immunohistochemical localization in human temporal lobe and induction by dimethylbenz(a)anthracene in astrocytoma cell line (MOG-G-CCM). Neurosci Lett 278:177–180
Salmena L, Poliseno L, Tay Y, Kats L, Pandolfi PP (2011) A ceRNA hypothesis: the rosetta stone of a hidden RNA language? Cell 146:353–358
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT et al (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504
Shaw EG, Wang M, Coons SW, Brachman DG, Buckner JC et al (2012) Randomized trial of radiation therapy plus procarbazine, lomustine, and vincristine chemotherapy for supratentorial adult low-grade glioma: initial results of RTOG 9802. J Clin Oncol 30:3065–3070
Son JC, Jeong HO, Park D, No SG, Lee EK et al (2017) miR-10a and miR-204 as a potential prognostic indicator in low-grade gliomas. Cancer Inform 16:1176935117702878
Sun G, Cao Y, Shi L, Sun L, Wang Y et al (2013) Overexpressed miRNA-137 inhibits human glioma cells growth by targeting Rac1. Cancer Biother Radiopharm 28:327–334
Ulitsky I, Bartel DP (2013) lincRNAs: genomics, evolution, and mechanisms. Cell 154:26–46
Wang DP, Tang XZ, Liang QK, Zeng XJ, Yang JB et al (2020) Overexpression of long noncoding RNA SLC26A4-AS1 inhibits the epithelial-mesenchymal transition via the MAPK pathway in papillary thyroid carcinoma. J Cell Physiol 235:2403–2413
Wang K, Jia Z, Zou J, Zhang A, Wang G et al (2013) Analysis of hsa-miR-30a-5p expression in human gliomas. Pathol Oncol Res 19:405–411
Wang X, Ding Y, Da B, Fei Y, Feng G (2018) Identification of potential prognostic long non-coding RNA signatures based on a competing endogenous RNA network in lung adenocarcinoma. Oncol Rep 40:3199–3212
Wang Y, Huang T, Sun X, Wang Y (2019) Identification of a potential prognostic lncRNA-miRNA-mRNA signature in endometrial cancer based on the competing endogenous RNA network. J Cell Biochem 120:18845–18853
Yao Y, Zhang T, Qi L, Liu R, Liu G et al (2019) Competitive endogenous RNA network construction and comparison of lung squamous cell carcinoma in smokers and nonsmokers. Dis Markers 2019:5292787
Yasin JK, Mishra BK, Arumugam Pillai M, Chinnusamy V (2021) Physical map of lncRNAs and lincRNAs linked with stress responsive miRs and genes network of pigeonpea (Cajanus cajan L.). J Plant Biochem Biotechnol
Yu X, Wu J, Hu M, Wu J, Zhu Q et al (2019) Glutamate affects the CYP1B1- and CYP2U1-mediated hydroxylation of arachidonic acid metabolism via astrocytic mGlu5 receptor. Int J Biochem Cell Biol 110:111–121
Zhang B, Wu Q, Xu R, Hu X, Sun Y et al (2019a) The promising novel biomarkers and candidate small molecule drugs in lower-grade glioma: evidence from bioinformatics analysis of high-throughput data. J Cell Biochem 120:15106–15118
Zhang F, Zhang L, Qi Y, Xu H (2016) Mitochondrial cAMP signaling. Cell Mol Life Sci 73:4577–4590
Zhang Y, Dong H, Duan L, Yuan G, Liang W et al (2019b) SLC1A2 mediates refractory temporal lobe epilepsy with an initial precipitating injury by targeting the glutamatergic synapse pathway. IUBMB Life 71:213–222
Zhengfeng Z, Huahua Z, Bo N, Benze X (2021) Theoretical and applied genetics
Ziegler N, Awwad K, Fisslthaler B, Reis M, Devraj K et al (2016) β-catenin is required for endothelial Cyp1b1 regulation influencing metabolic barrier function. J Neurosci 36:8921–8935
Acknowledgements
YM Ding and HJ Liu performed the data analyses and wrote the manuscript. ZS Bao and CB Zhang contributed significantly to manuscript revision. SQ Yu conceived and designed the study. All authors read and approved the final manuscript and agreed to be accountable for all aspects of the research in ensuring that the accuracy or integrity of any part of the work are appropriately investigated and resolved. We acknowledge TCGA database for providing their platforms and contributors for uploading their meaningful datasets.
Funding
This study was supported by the General Technological Program of the Peking Education Commission (KM201810025023).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Ethical approval
There was no ethical review or informed consent required.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Ding, Y., Liu, H., Zhang, C. et al. Comprehensive analysis of the LncRNAs, MiRNAs, and MRNAs acting within the competing endogenous RNA network of LGG. Genetica 150, 41–50 (2022). https://doi.org/10.1007/s10709-021-00145-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-021-00145-3