Abstract
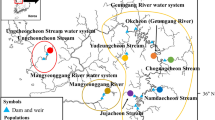
Recently, anthropogenic alterations have had severe and negative impacts on the terrestrial and aquatic species and environments. To conserve species that have a small and limited habitat, it is necessary to focus on fine-scale population structure and its effects on persistence. The deepbodied bitterling Acheilognathus longipinnis is an endangered freshwater fish that occupies ponds scattered in lateral bars in the Kiso River. In this study, we conducted multi-locus microsatellite DNA analysis to evaluate both fine-scale population structure and genetic diversity, in order to conserve A. longipinnis. The smaller number of loci deviating from the Hardy–Weinberg equilibrium in ponds scattered in individual lateral bars compared to the whole river system suggests that A. longipinnis forms a local breeding population in units of ponds. The population was roughly split between the river banks and the local population located in ponds in the mid-channel bar showed intermediate relationships with the river bank populations. Gene flow between local populations was not always homogeneous and was not influenced by geographical distances between local populations or the direction of river flow. The dispersal of A. longipinnis across both river bank sides may be constrained and is probably affected by the ecological characteristics of A. longipinnis and the hydrological regimes. Consequently, A. longipinnis in the Kiso River is maintained as a complex of multiple local populations with appropriate gene flow among them. To conserve A. longipinnis, both the persistence of the unstable ponds and moderate genetic exchanges by individual migration are required.



Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author, YY, upon reasonable request.
Code availability
Not applicable.
References
Avise JC (2000) Phylogeography: the history and formation of species. Harvard University Press, Cambridge
Barson NJ, Cable J, van Oosterhout C (2009) Population genetic analysis of microsatellite variation of guppies (Poecilia reticulata) in Trinidad and Tobago: evidence for a dynamic source-sink metapopulation structure, founder events and population bottlenecks. J Evol Biol 22:485–497
Bartáková V, Bryja J, Reichard M (2018) Fine-scale genetic structure of the European bitterling at the intersection of three major European watersheds. BMC Evol Biol 18:105
Bartáková V, Reichard M, Blažek R, Polačik M, Bryja J (2015) Terrestrial fishes; rivers are barriers to gene flow in annual fishes from the African savanna. J Biogeogr 42:1832–1844
Belkhir K, Borsa P, Chikhi N, Raufaste N, Bonhomme F (1996–2004) GENETIX 4.05, Logiciel Sous WindowsTM Pour la Génétique des Populations. Laboratoire Genome, Populations, Interactions, CNRS UMR 5000, Université de Montpellier II, Montpellier
Benton TG, Vickery JA, Wilson JD (2003) Farmland biodiversity: is habitat heterogeneity the key? Trends Ecol Evol 18:182–188
Bergek S, Olsson J (2009) Spatiotemporal analysis shows stable genetic differentiation and barriers to dispersal in the Eurasian perch (Perca fluviatilis L.). Evol Ecol Res 11:827–840
Carlsson J, Olsén KH, Nilsson J, Øverli Ø, Stabell OB (1999) Microsatellite reveal fine-scale genetic structure in stream-living brown trout. J Fish Biol 55:1290–1303
Castric V, Bernatchez L, Belkhir K, Bonhomme F (2002) Heterozygote deficiencies in small lacustrine populations of brook charr Salvelinus fontinalis Mitchill (Pisces, Salmonidae): a test of alternative hypotheses. Heredity 89:27–35
Cook BD, Kennard MJ, Adams M, Raadik TA, Real K, Bunn SE, Hughes JM (2019) Hydrographic correlates of within-river distribution and population genetic structure in two widespread species of mountain galaxias (Teleostei, Galaxiidae) in southern Australia. Freshw Biol 64:506–519
Cornuet JM, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2004
Dawson DA, Burland TM, Douglas A, Le Comber SC, Bradshaw M (2003) Isolation of microsatellite loci in the freshwater fish, the bitterling Rhodeus sericeus (Teleostei: Cyprinidae). Mol Ecol Notes 3:199–202
Earl DA, vonHoldt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer HEL (2010) ARLEQUIN suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Excoffier L, Smouse PE, Quatrro JM (1992) Analysis of molecular variance inferred from metric distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Ferreira DG, Lima SC, Frantine-Silva W, Silva JF, Apolinário-Silva C, Sofia SH, Carvalho S, Galindo BA (2016) Fine-scale genetic structure patterns in two freshwater fish species, Geophagus brasiliensis (Osteichthyes, Cichlidae) and Astyanax altiparanae (Osteichthyes, Characidae) throughout a Neotropical stream. Genet Mol Res 15:gmr15048124
Ferreira DG, Souza-Shibatta L, Shibatta OA, Sofia SH, Carlsson J, Dias JHP, Makrakis S, Makrakis MC (2017) Genetic structure and diversity of migratory freshwater fish in a fragmented Neotropical river system. Rev Fish Biol Fish 27:209–231
Frankham R (1996) Relationship of genetic variation to population size in wildlife. Conserv Biol 10:1500–1508
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Frantz AC, Bertouille S, Eloy MC, Licoppe A, Chaumont F, Flamand MC (2012) Comparative landscape genetic analyses show a Belgian motorway to be a gene flow barrier for red deer (Cervus elaphus), but not wild boars (Sus scrofa). Mol Ecol 21:3445–3457
Futuyma DJ (1998) Evolutionary biology, 3rd edn. Sinauer Associates, Inc., Sunderland
Hanski I (1998) Metapopulation dynamics. Nature 396:41–49
Hardy OJ, Vekemans X (2002) SPAGeDi: a versatile computer program to analyze spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hasegawa K, Kanao S, Miyazaki Y, Mukai T, Nakajima J, Takaku K, Taniguchi Y (2019) Acheilognathus longipinnis. The IUCN Red List of Threatened Species 2019. http://www.iucnredlist.org. Accessed 7 July 2020
Hughes JM, Schmidt DJ, Finn DS (2009) Genes in streams: using DNA to understand the movement of freshwater fauna and their riverine habitat. Bioscience 59:573–583
Hurt C, Kuhajda B, Harman A, Ellis N, Nalan M (2017) Genetic diversity and population structure in the Barrens Topminnow (Fundulus julisia): implications for conservation and management of a critically endangered species. Conserv Genet 18:1347–1358
Ikeya K, Sagawa S, Ohara K (2012) The efforts of the ex situ conservation of endangered deep body bitterling in Nobi-Plain, Japan. Reintroduction 2:121–128 (in Japanese)
Ito T, Tominaga A (2016) A study on generation of scour due to streams around large-scale groins under tidal effect. J Jpn Soc Civ Eng B1(72):829–834 (in Japanese with English Abstract)
IUCN (2008) Wildlife in a changing world: an analysis of the 2008 IUCN Red List of Threatened Species.
Japan Ministry of the Environment (2003) Threatened wildlife of Japan, red data book, 2nd edn. Japan Wildlife Research Center, Tokyo (in Japanese)
Jeon HB, An J, Kweon SM, Kim S, Yu JN, Kim BJ, Kawase S, Suk HY (2016) Development of novel microsatellite loci and analyses of genetic diversity in the endangered Tanakia somjinensis. Biochem Syst Ecol 66:344–350
Johnson MS, Black R (1984) The Wahlund effect and the geographical scale of variation in the intertidal limpet Siphonaria sp. Mar Biol 79:295–302
Kalinowski ST (2005) HP-Rare: a computer program for performing rarefaction on measures of allelic diversity. Mol Ecol Notes 5:187–189
Kamibayashi Y (1993) On the social background of Kisogawa River improvement in the dawn of Meiji Japan, planed by J. de Rijke using modern civil engineering. Hist Stud Civ Eng 13:375–386 (in Japanese with English Abstract)
Kanno Y, Vokoun JC, Letcher BH (2011) Fine-scale population structure and riverscape genetics of brook trout (Salvelinus fortinalis) distributed continuously along head water channel networks. Mol Ecol 20:3711–3729
Karlsson S, Mork J (2005) Deviation from Hardy-Weinberg equilibrium, and temporal instability in allele frequencies at microsatellite loci in a local population of Atlantic cod. ICES J Mar Sci 62:1588–1596
Kitanishi S, Nishio M, Sagawa S, Uehara K, Ogawa R, Yokoyama T, Ikeya K, Edo K (2013) Strong population genetic structure and its implications for the conservation and management of the endangered Itasenpara bitterling. Conserv Genet 14:901–906
Kopelman NM, Mayzel Z, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Res 15:1179–1191
Kume M, Onoda Y, Negishi JN, Sagawa S, Nagayama S, Kayaba Y (2012) Feeding damage by exotic species, nutria (Myocastor coypus), to unionid mussels in a floodplain water-body of the Kiso River, Japan. Biol Inland Waters 27:41–47
Lasne E, Lek S, Laffaille P (2007) Patterns in fish assemblages in the Loire floodplain: the role of hydrological connectivity and implications for conservation. Biol Conserv 139:258–268
Lord JM, Norton DA (1990) Scale and the spatial concept of fragmentation. Conserv Biol 4:197–202
Luikart G, Allendorf FW, Cornuet J-M, Sherwin WB (1988) Distribution of allele frequency distributions provide a test for recent population bottlenecks. J Hered 89:238–247
Lynch M, Ritland K (1999) Estimation of pairwise relatedness with molecular markers. Genetics 152:1753–1766
Marsh DM, Trenham PC (2001) Metapopulation dynamics and amphibian conservation. Conserv Biol 15:40–49
Matsuura S, Fujii M (1993) A study on the administration of river works at the beginning of Meiji era. Hist Stud Civ Eng 13:145–160 ((in Japanese))
Monastersky R (2014) Biodiversity: life—a status report. Nature 516:158–161
Morimoto J, Yoshida H (2005) Dynamic changes of native Rhododendron colonies in the urban fringe of Kyoto City in Japan: detecting the long-term dynamism for conservation of secondary nature. Landsc Urban Plan 70:195–204
Moritz C (1994) Defining ‘Evolutionarily Significant Units’ for conservation. Trends Ecol Evol 9:373–375
Nagayama S, Harada M, Kayaba Y (2015) Can floodplains be recovered by flood-channel excavation? An example from Japanese lowland rivers. Ecol Civ Eng 17:67–77 (in Japanese with English Abstract)
Narum SR (2006) Beyond Bonferroni: less conservative analyses for conservation genetics. Conserv Genet 7:783–787
Nishio M, Kawamoto T, Kawakami R, Edo K, Yamazaki Y (2015) Life history and reproductive ecology of the endangered Itasenpara bitterling Acheilognathus longipinnis (Cyprinidae) in the Himi region, central Japan. J Fish Biol 87:616–633
Nishio M, Tanaka H, Tanaka D, Kawakami R, Edo K, Yamazaki Y (2016) Managing water levels in rice paddies to conserve the Itasenpara Host Mussel, Unio douglasiae nipponensis. J Shellfish Res 35:1–7
Ogawa R (2008) Acheilognathus longipinnis: a symbol fish of flood plains with natural hydrometeorological environments. Jpn J Ichthyol 55:144–148 (in Japanese with English Abstract)
Okazaki T, Watanabe M, Inamura O, Kitagawa T, Tabe M, Nagata Y (2006) Genetic relationships among regional populations of the deepbodied bitterling, Acheilognathus longipinnis, inferred from mitochondrial DNA analysis. DNA Polymorph 14:276–280 (in Japanese)
Piry S, Luikart G, Cornuet JM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 153:945–959
Rice WR (1989) Analyzing tables of statistical test. Evolution 43:223–225
Richmond JQ, Backlin AR, Galst-Cavalcante C, O’Brien JW, Fisher RN (2018) Loss of dendritic connectivity in southern California’s urban riverscape facilitates decline of an endemic freshwater fish. Mol Ecol 27:369–386
Richter BD, Baumgartner JV, Braun DP, Powell J (1998) A spatial assessment of hydrologic alteration within a river network. Regul Rivers Res Manag 14:329–340
Sanjou M, Goto S, Oishi T, Takaoka H, Okamoto T (2017) Fundamental study on dissolved oxygen transport from mainstream to Wando zone. J Jpn Soc Civ Eng B1(73):739–744 (in Japanese with English Abstract)
Shirai Y, Ikeda S, Tajima S (2009) Isolation and characterization of new microsatellite markers for rose bitterlings, Rhodeus ocellatus. Mol Ecol Res 9:1031–1033
Takaoka H, Nagayama S, Kayaba Y (2014) Investigation of river-bed morphology and generating process of local scouring in the Kiso River. J Jpn Soc Civ Eng B1(70):1015–1020 (in Japanese with English Abstract)
Takeda M, Kimura I, Matsuo N, Yamazaki Y, Fujita A (2002) Study on water temperature change and its numerical simulation model in Kiso River Wando. Annu J Hydraul Eng JSCE 46:1097–1102 (in Japanese with English Abstract)
Tilman D, Fargione J, Wolff B, D’Antonio C, Dobson A, Howarth R, Schindler D, Schlesinger WH, Simberloff D, Swackhamer D (2001) Forecasting agriculturally driven global environmental change. Science 292:281–284
Tockner K, Stanford JA (2002) Riverine flood plains: present state and future trends. Environ Conserv 29:308–330
van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Wang J (2012) Computationally efficient sibship and parentage assignment from multilocus marker data. Genetics 191:183–194
Waples RS (1998) Separating the wheat from the chaff: patterns of genetic differentiation in high gene flow species. J Hered 89:438–450
Ward JV, Tockner K, Schiemer F (1999) Biodiversity of floodplain river ecosystems: ecotones and connectivity. Regul Rivers Res Manag 15:125–139
Wilson GA, Rannala B (2003) Bayesian inference of recent migration rates using multilocus genotypes. Genetics 163:1177–1191
Yamamoto S, Morita K, Koizumi I, Maekawa K (2004) Genetic differentiation of white-spotted charr (Salvelinus leucomaenis) populations after habitat fragmentation: spatial–temporal changes in gene frequencies. Conserv Genet 5:529–538
Yamazaki Y, Nakamura T, Nishio M (2010) Genetic population structures of the Itasenpara bitterling, Acheilognathus longipinnis, in the Toyama and Osaka regions. Jpn J Ichthyol 57:143–148 (in Japanese with English Abstract)
Yamazaki Y, Yamano A, Oura K (2011) Recent microscale disturbance of gene flow in threatened fluvial lamprey, Lethenteron sp. N, living in a paddy water system. Conserv Genet 12:1373–1377
Yamazaki Y, Yokoyama R, Nagai T, Goto A (2014a) Population structure and gene flow among anadromous arctic lamprey (Lethenteron camtschaticum) populations deduced from polymorphic microsatellite loci. Environ Biol Fish 97:43–52
Yamazaki Y, Nakamura T, Sasaki M, Nakano S, Nishio M (2014b) Decreasing genetic diversity in wild and captive populations of endangered Itasenpara bitterling (Acheilognathus longipinnis) in the Himi region, central Japan, and recommendations for conservation. Conserv Genet 15:921–932
Yamazaki Y, Ikeya K, Goto T, Chimura Y (2017) Population viability analysis predicts decreasing genetic diversity in ex-situ populations of the Itasenpara bitterling Acheilognathus longipinnis from the Kiso River, Japan. Ichthyol Res 64:54–63
Yamazaki Y, Uehara K, Ikea K, Nishio M (2020) Interpopulational and intrapopulational genetic diversity of the endangered Itasenpara bitterling (Acheilognathus longipinnis) with reference to its demographic history. Conserv Genet 21:55–64
Yokoyama R, Yamano A, Takeshima H, Nishida M, Yamazaki Y (2009) Disturbance of the indigenous gene pool of the threatened brook lamprey Lethenteron sp. S by intraspecific introgression and habitat fragmentation. Conserv Genet 10:29–43
Yoshii T, Ozaki M (1986) History and perspective of water resources development in the Yodo River basin. J Jpn Soc Irrig Drain Reclam 54:603–610
Acknowledgements
We thank Masayuki Kato, Ryota Noguchi, and the other members of Environmental Conservation Network, for collecting samples. We are also grateful to the Conservation Council for Itasenpara (endangered bitterling) of Kiso River System, Ministry of Land, Infrastructure, Transport and Tourism (MLIT), Ministry of the Environment of Japan, and municipal governments. This research was carried out as part of the program for Rehabilitation of Natural Habitats and Maintenance of Viable Population for Itasenpara bitterling (Acheilognathus longipinnis) by Chubu Regional Environment Office, Ministry of the Environment. A part of this study was supported by the research fund for the Kiso River provided by the MLIT of Japan to S. M. We would like to thank Editage (www.editage.com) for English language editing.
Author information
Authors and Affiliations
Contributions
YY completed laboratory work and data analysis, performed the research, and wrote the manuscript. JK conducted sample collection and edited the manuscript. KI conducted sample collection and edited the manuscript. SM corresponded with the administrative agency regarding collection permission, and edited the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no competing interests.
Ethical approval
This study was conducted with respect for the conservation of the target endangered species and its habitats. The DNA sample used was a scale extracted from an individual, and the individual was released alive in its habitat.
Informed consent
Not applicable.
Consent to publish
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10709_2021_123_MOESM1_ESM.pptx
Supplementary file1 (PPTX 54 kb) Fig. S1 The values of mean LnP (K) (A), ΔK (B), and Prb (K) (C) for K in eight local populations of Acheilognathus longipinnis in the Kiso River
10709_2021_123_MOESM2_ESM.pptx
Supplementary file2 (PPTX 84 kb) Fig. S2 Bar plot showing Bayesian assignment of Acheilognathus longipinnis individuals in the Kiso River estimated by STRUCTURE software with K = 2 and 3. Each bar represents the estimated admixture coefficient (q) for each individual in each cluster
Rights and permissions
About this article
Cite this article
Yamazaki, Y., Kitamura, ·i., Ikeya, K. et al. Fine-scale genetic structure of the endangered bitterling in the middle river basin of the Kiso River, Japan. Genetica 149, 179–190 (2021). https://doi.org/10.1007/s10709-021-00123-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-021-00123-9