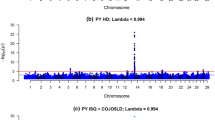
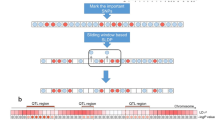
Abstract
SNP chips are commonly used for genotyping animals in genomic selection but strategies for selecting low-density (LD) SNPs for imputation-mediated genomic selection have not been addressed adequately. The main purpose of the present study was to compare the performance of eight LD (6K) SNP panels, each selected by a different strategy exploiting a combination of three major factors: evenly-spaced SNPs, increased minor allele frequencies, and SNP-trait associations either for single traits independently or for all the three traits jointly. The imputation accuracies from 6K to 80K SNP genotypes were between 96.2 and 98.2%. Genomic prediction accuracies obtained using imputed 80K genotypes were between 0.817 and 0.821 for daughter pregnancy rate, between 0.838 and 0.844 for fat yield, and between 0.850 and 0.863 for milk yield. The two SNP panels optimized on the three major factors had the highest genomic prediction accuracy (0.821–0.863), and these accuracies were very close to those obtained using observed 80K genotypes (0.825–0.868). Further exploration of the underlying relationships showed that genomic prediction accuracies did not respond linearly to imputation accuracies, but were significantly affected by genotype (imputation) errors of SNPs in association with the traits to be predicted. SNPs optimal for map coverage and MAF were favorable for obtaining accurate imputation of genotypes whereas trait-associated SNPs improved genomic prediction accuracies. Thus, optimal LD SNP panels were the ones that combined both strengths. The present results have practical implications on the design of LD SNP chips for imputation-enabled genomic prediction.






Similar content being viewed by others
Abbreviations
- ANOVA:
-
Analysis of variance
- DPR:
-
Daughter pregnancy rate
- FY:
-
Fat yield
- GEBV:
-
Genomic-estimated breeding value
- GER:
-
Genotype (imputation) error rate
- GPA:
-
Genomic prediction accuracy
- GS:
-
Genomic selection
- HD:
-
High-density
- LD:
-
Low-density
- LGPA:
-
Loss in genomic prediction accuracy
- MAF:
-
Minor allele frequencies
- MCMC:
-
Markov chain Monte Carlo
- MD:
-
Moderate-density
- MOLO:
-
Multiple-objective, local-optimization
- MY:
-
Milk yield
- PTAs:
-
Predicted transmitting abilities
- RGPA:
-
Relative genomic prediction accuracy
- RTMGL:
-
Relative total maximum gap length
- TMGL:
-
Total maximum gap length
References
Boichard D, Chung H, Dassonneville R, David X, Eggen A, Fritz S, Van Tassell CP (2012) Design of a bovine low-density SNP array optimized for imputation. PloS one 7:e34130
Bolormaa S, Gore K, Werf JHJ, Hayes BJ, Daetwyler HD (2015) Design of a low density SNP chip for the main Australian sheep breeds and its effect on imputation and genomic prediction accuracy. Anim Genet 46:544–556
Calus MPL, Bouwman AC, Hickey JM, Veerkamp RF, Mulder HA (2014) Evaluation of measures of correctness of genotype imputation in the context of genomic prediction: a review of livestock applications. Animal 8:1743–1753
Cooper TA, Wiggans GR, VanRaden PM (2015) Short communication: analysis of genomic predictor population for Holstein dairy cattle in the United States—effects of sex and age. J Dairy Sci 98:2785–2788
Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH (2010) Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet 11:446
Erbe M, Hayes BJ, Matukumalli LK, Goswami S, Bowman PJ, Reich CM. Mason BA, Goddard ME (2012) Improving accuracy of genomic predictions within and between dairy cattle breeds with imputed high-density single nucleotide polymorphism panels. J Dairy Sci 95:4114–4129
Habier D, Fernando RL, Dekkers JCM (2009) Genomic selection using LD marker panels. Genetics 182:343–353
Habier D, Fernando RL, Kizilkaya K, Garrick DJ (2011) Extension of the bayesian alphabet for genomic selection. BMC Bioinform 12:186
Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Jia Y, Jannink JL (2012) Multiple-trait genomic selection methods increase genetic value prediction accuracy. Genetics 192:1513–1522
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. IJCAI-1995. Morgan Kaufmann, San Mateo. 2:pp 1137–1143
Manolio TA, Collins FS, Cox NJ, Goldstein DB, Hindorff LA, Hunter DJ, McCarthy MI, Ramos EM, Cardon LR, Chakravarti A, Cho JH, Guttmacher AE, Kong A, Kruglyak L, Mardis E, Rotimi CN, Slatkin M, Valle D, Whittemore AS, Boehnke M, Clark AG, Eichler EE, Gibson G, Haines JL, MacKay TFC, McCarroll SA, Visscher PM (2009) Finding the missing heritability of complex diseases. Nature 461:747–753
Pimentel EC, Wensch-Dorendorf M, König S, Swalve HH (2013) Enlarging a training set for genomic selection by imputation of ungenotyped animals in populations of varying genetic architecture. Genet Sel Evol 45:12
Sargolzaei M, Chesnais JP, Schenkel FS (2014) A new approach for efficient genotype imputation using information from relatives. BMC Genom 15:478
van der Werf J (2013) Genomic selection in animal breeding programs. In: Gondro C, van der Werf J, Hayes BJ (ed) Genome-wide association studies and genomic prediction. Springer, New York, pp 543–561
VanRaden PM, Van Tassell CP, Wiggans GR, Sonstegard TS, Schnabel RD, Taylor JF, Schenkel FS (2009) Invited review: reliability of genomic predictions for North American Holstein bulls. J Dairy Sci 92:16–24
Weigel KA, de los Campos G, Gonzalez O. Naya H, Wu XL, Long N, Rosa GJM, Gianola D (2009) Predicting ability of direct genomic values for lifetime net merit of Holstein sires using selected subsets of single nucleotide polymorphism markers. J Dairy Sci 92:5248–5257
Wiggans GR, Sonstegard TS, VanRaden PM, Matukumalli LK, Schnabel RD, Taylor JF, Schenkel FS, Van Tassell CP (2009) Selection of single-nucleotide polymorphisms and quality of genotypes used in genomic evaluation of dairy cattle in the United States and Canada. J Dairy Sci 92:3431–3436
Wiggans GR, Cooper TA, Vanraden PM, Olson KM, Tooker ME (2012) Use of the Illumina Bovine3K BeadChip in dairy genomic evaluation. J Dairy Sci 95:1552–1558
Wu XL, Sun C, Beissinger TM, Rosa GJ, Weigel KA, Gatti Nde L, Gianola D (2012a) Parallel Markov chain Monte Carlo bridging the gap to high-performance Bayesian computation in animal breeding and genetics. Genet Sel Evol 44:29
Wu XL, Hayrettin O, Duan H, Beissinger T, Bauck S, Woodward B, Rosa GJ, Weigel KA, de Leon Gatti N, Taylor J, Gianola D (2012b) Parallel-BayesCpC on OSG: grid-enabled high-throughput computing for genomic selection in practice. PAG XX, San Diego
Wu XL, Xu J, Feng G, Wiggans GR, Taylor JF, He J, Qian C, Qiu J, Simpson B, Walker J, Bauck S (2016) Optimal design of low-density SNP arrays for genomic prediction: algorithm and applications. PLoS ONE 11:e0161719
Zuk O, Hechter E, Sunyaev SR, Lander ES (2012) The mystery of missing heritability: genetic interactions create phantom heritability. PNAS 109:1193–1198
Author information
Authors and Affiliations
Contributions
JH and JX analyzed the data. JH and XW drafted the manuscript. XW,JL,SB,GM,SK and MS participated in it’s the design and discussions of this research. All authors have proof-read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interests in this work.
Funding
JH, JX, and JL acknowledge the financial support by University of Nebraska–Lincoln, and GeneSeek (A Neogen company). HJ was also supported by the Bairen Plan of Hunan Province, China (XZ2016-08-07) and Hunan Co-Innovation center of Animal Production Safety, China.
Additional information
Jun He and Jiaqi Xu contributed equally.
Rights and permissions
About this article
Cite this article
He, J., Xu, J., Wu, XL. et al. Comparing strategies for selection of low-density SNPs for imputation-mediated genomic prediction in U. S. Holsteins. Genetica 146, 137–149 (2018). https://doi.org/10.1007/s10709-017-0004-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-017-0004-9