Abstract
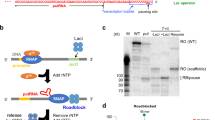
Eukaryotic RNA polymerase III (RNAP III) transcribes tRNA genes and short interspersed elements that have internal promoters consisting of A- and B-blocks. The B-block binding subunit of the transcription initiation factor TFIIIC binds to the B-block. The mobile bacterial insertion sequence (IS) 1 contains a RNAP III promoter-like sequence, which stimulates bacterial transcription along with the bacterial ArtA protein. Here, the DNA-binding ability of ArtA was examined in vitro using a simple, newly developed method. Various DNA fragments, including RNAP III promoter fragments, were separately incubated with purified ArtA, and then loaded onto a polyacrylamide gel. Since DNAs bound by ArtA remain in the gel wells during electrophoresis, SDS was added into the wells at the electrophoresis halfway point. It was hypothesized that SDS would dissociate the DNA–ArtA complexes in the wells, and then the DNAs would begin to migrate. In fact, new bands appeared in all of the lanes at similar intensities, indicating that ArtA binds nonspecifically to DNA. Therefore, labeled wild-type RNAP III promoter fragments were incubated with either the unlabeled wild-type or mutant fragments and ArtA, and electrophoresed. The B-block(-like) sequences of IS1, a human Alu element, and an anuran tRNA gene were important for binding to ArtA. Additionally, in silico analyses revealed the presence of the RNAP III promoter-like structures in the IS1 isoforms and the IS3 family elements. These results suggest the presence of parts of the RNAP III transcription machinery in bacteria, and might imply that its prototype existed in the common ancestor.






Similar content being viewed by others
References
Archambault J, Friesen JD (1993) Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol Rev 57:703–724
Bartholomew B, Kassavetis GA, Braun BR, Geiduschek EP (1990) The subunit structure of Saccharomyces cerevisiae transcription factor IIIC probed with a novel photocrosslinking reagent. EMBO J 9:2197–2205
Blombach F, Makarova KS, Marrero J, Siebers B, Koonin EV, van der Oost J (2009) Identification of an ortholog of the eukaryotic RNA polymerase III subunit RPC34 in Crenarchaeota and Thaumarchaeota suggests specialization of RNA polymerases for coding and non-coding RNAs in Archaea. Biol Direct 4:39
Bondos SE, Bicknell A (2003) Detection and prevention of protein aggregation before, during, and after purification. Anal Biochem 316:223–231
Brun I, Sentenac A, Werner M (1997) Dual role of the C34 subunit of RNA polymerase III in transcription initiation. EMBO J 16(18):5730–5741
Galli G, Hofstetter H, Birnstiel ML (1981) Two conserved sequence blocks within eukaryotic tRNA genes are major promoter elements. Nature 294:626–631
Hirata A, Murakami KS (2009) Archaeal RNA polymerase. Curr Opin Struct Biol 19:724–731
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9
Macrina FL, Jones KR, Laloi P (1996) Characterization of IS199 from Streptococcus mutans V403. Plasmid 36:9–18
Mahillon J, Chandler M (1998) Insertion sequences. Microbiol Mol Biol Rev 2:725–774
Matsutani S (2004) Similarities in transcription factor IIIC subunits that bind to the posterior regions of internal promoters for RNA polymerase III. BMC Evol Biol 4:26
Matsutani S (2005) The internal sequence of IS1 stimulates RNA synthesis from the IS1 own and exogenous promoters. J Biol Syst 13:313–329
Matsutani S (2007) Possible presence and role of the promoter sequence for eukaryotic RNA polymerase III in bacteria. Genetica 131:127–134
Matsutani S (2012) Bacterial ArtA protein specifically binds to the internal region of IS1 in vitro. Adv Biosci Biotechnol 3:869–875
Matsutani S (2014) Evolution of the B-block binding subunit of TFIIIC that binds to the internal promoter for RNA Polymerase III. Int J Evol Biol 2014:609865
Murakami KS, Darst SA (2003) Bacterial RNA polymerases: the wholo story. Curr Opin Struct Biol 13:31–39
Ohtsubo E, Ohtsubo H, Doroszkiewicz W, Nyman K, Allen D, Davison D (1984) An evolutionary analysis of iso-IS1 elements from Escherichia coli and Shigella strains. J Gen Appl Microbiol 30:359–376
Perez-Stable C, Ayres TM, Shen C-KJ (1984) Distinctive sequence organization and functional programming of an Alu repeat promoter. Proc Natl Acad Sci USA 81:5291–5295
Rasband WS (1997–2015) ImageJ. http://imagej.nihs.gov/ij/
Sambrook J, Fritsch EF, Maniatis T (1989) Molecular coloning. Cold Spring Harbor Laboratory Press, New York
Schramm L, Hernandez N (2002) Recruitment of RNA polymerase III to its target promoters. Genes Dev 16:2593–2620
Shimizu-Kadota M, Kiwaki M, Hirokawa H, Tsuchida N (1985) ISL1: a new transposable element in Lactobacillus casei. Mol Gen Genet 200:193–198
Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Söding J, Thompson JD, Higgins DG (2011) Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 7:539
White RJ (2011) Transcription by RNA polymerase III: more complex than we thought. Nat Rev Genet 12:459–463
Wu JH, Ippen-Ihler K (1989) Nucleotide sequence of traQ and adjacent loci in the Escherichia coli K-12 F-plasmid transfer operon. J Bacteriol 171:213–221
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10709_2016_9905_MOESM1_ESM.pptx
Supplementary Fig. 1 ImageJ plots of the gel images in Fig. 3. Boxed regions were plotted in the directions of arrows, and the areas under the peaks were measured. All of the areas are shown as the ratios to the area of the peak at the left end (or the upper left) in each of the plot figures. The average values from independent experiments are shown in brackets below the plot figures. The original raw images were used for quantification of the fluorescent probe signals. A IS1 fragment containing the RNAP III promoter-like sequence. 0.3 pmol of the 6FAM-labeled IS1AB361was loaded to the marker lanes while reaction mixtures containing 1.5 pmol of the probe were loaded to the assay sample lanes. B Human Alu RNAP III promoter fragment. The image was cropped to show only the lanes with competitor DNAs. C RNAP III promoter fragment of the X. laevis tRNALue gene. The image was cropped to show only the lanes with competitor DNAs. D Quantified binding activity of ArtA to each of the wild-type and mutant RNAP III promoters of the Alu element (PPTX 2225 kb)
10709_2016_9905_MOESM2_ESM.pptx
Supplementary Fig. 2 The Clustal Omega alignment of nucleotide sequences of the elements belonging to the IS3 group (PPTX 64 kb)
10709_2016_9905_MOESM3_ESM.pptx
Supplementary Fig. 3 The Clustal Omega alignment of nucleotide sequences of the elements belonging to the IS51 group and IS600. IS1372 is at bp 58–1361 in U50076, and IS401 is at bp 130–1445 in L09108 (PPTX 74 kb)
10709_2016_9905_MOESM4_ESM.pptx
Supplementary Fig. 4 The Clustal Omega alignment of nucleotide sequences of the elements belonging to the IS407 group and IS1236 (PPTX 75 kb)
Rights and permissions
About this article
Cite this article
Matsutani, S. Possible interaction between the bacterial transcription factor ArtA and the eukaryotic RNA polymerase III promoter. Genetica 144, 361–374 (2016). https://doi.org/10.1007/s10709-016-9905-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-016-9905-2