Abstract
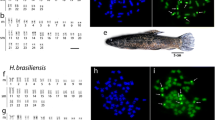
This paper describes the karyotype analysis of Haemulon aurolineatum, Haemulon bonariensis and Haemulon plumierii, by Giemsa staining, C-banding, Ag-staining and fluorescent in situ hybridization (FISH), to locate the 18S and 5S rRNA genes. Diploid modal count in the three species was 2n = 48 acrocentric elements. Except for pair 24, which exhibited an unmistakable secondary constriction in all three species, it was not possible to classify them as homologous to each other because differences in chromosome size were too slight between adjacent pairs within a size-graded series. Ag-NOR clusters were located in pair 24 in the three species with signal located on the secondary constriction of these chromosomes. C-banding demonstrated that the three species share the same distribution pattern of the constitutive heterochromatin with centromeric heterochromatic blocks in the 23 chromosome pairs and a pericentromeric block in pair 24 which is coincident with the NORs. FISH experiments showed that 18S rDNA sequences were located coincident with the Ag-NOR site in the three species; however, differences in both the number and chromosome distribution of 5S-rDNA cluster were detected among them. Our data suggest that chromosome evolution of Haemulon has been preserved from major changes in the karyotypic macrostructure, whereas microstructural changes have occurred.



Similar content being viewed by others
References
Accioly IV, Molina WF (2004) Contribuição à citogenética dos gêneros Pomadasys e Anisotremus (Haemulidae, Perciformes). X Simpósio de Citogenética e Genética de Peixes. Universidade Federal do Rio Grande do Norte, Natal, Rio Grande do Norte, Brazil
Appeldoorn RS, Lindeman KC (1985) Multispecies assessment in coral reef fisheries using higher taxonomic categories as unit stocks, with an analysis of an artisanal haemulid fishery. In: Proceedings of the 5th International Coral Reef Congress, Tahiti 27 May–1 June 1985, vol 5. Miscellaneous papers (A), pp 507–514
Brum MJI (1996) Cytogenetic studies of Brazilian marine fish. Brazil J Genet 19(3):421–427
Brum MJI, Galetti PM (1997) Teleostei ground plan karyotype. J Comp Biol 2(2):91–102
Cequea H, Pérez JE (1971) Variación intra e interespecífica de hemoglobina y proteínas del plasma en algunas especies del género Haemulon. Bol Inst Oceanograf Univ Oriente 10(2):79–85
Cervigón F (1993) Los peces marinos de Venezuela, 2nd ed, vol 2. Fundación Científica Los Roques, Caracas, Venezuela
Duran GA, García CE, Laguarda A (1990) The karyotype and “G” bands of Haemulon aurolienatum Cuvier, 1829 (Pisces: Haemulidae). An Inst Cienc Mar Limnol Univ Nac Auton Mex 17(2):299–307
Foresti F, Oliveira C, Almeida-Toledo LF (1993) A method for chromosome preparations from large specimens of fishes using in vitro short treatment with colchicine. Experientia 49:810–813
Garcia E, Alvarez MC, Thode G (1987) Chromosome relationships in the genus Blennius (Blennidae, Perciformes). C-banding patterns suggest two karyoevolutional pathways. Genetica 72:27–36
Gornung E, Gabrielli I, Cataudella S, Sola L (1997) CMA3-banding pattern and fluorescence in situ hybridization with 18S rDNA genes in zebrafish chromosomes. Chromosome Res 5:40–46
Gromicho M, Ozouf-Costaz C, Collares-Pereira MJ (2005) Lack of correspondence between CMA3-, Ag-positive signals and 28S rDNA loci in two Iberian minnows (Teleostei, Cyprinidae) evidenced by sequential banding. Cytogenet Genome Res 109:507–511
Howell WM, Black DA (1980) Controlled silver-staining of nucleolus organizer regions with a protective colloidal developer: a 1-step method. Experientia 36:1014–1015
Levan A, Fredga K, Sandberg A (1964) Nomenclature for centrometric position on chromosomes. Hereditas 52:201–220
Lima LCB, Molina WF (2004) Homeostase cariotípica em três espécies do gênero Haemulon (Haemulidae, Perciformes) do litoral Potiguar. X Simpósio de Citogenética e Genética de Peixes. Universidade Federal do Rio Grande do Norte, Natal, Rio Grande do Norte, Brazil
Lindeman KC (2002) Haemulidae (Grunts). In: Carpenter KE (ed) The living marine resources of the Western Central Atlantic, vol 3. Bony fishes. Part 2 (Opistognathidae to Molidae), sea turtles and marine mammals. FAO Species Identification Guide for Fishery Purposes and American Society of Ichthyologists and Herpetologists Special Publication No. 5. Rome, FAO. 2002. pp 1522–1529
Lucchini S, Nardi I, Barsacchi G, Batistoni R, Andronico F (1993) Molecular cytogenetics of the ribosomal (18S + 28S and 5S) DNA loci in primitive and advanced urodele amphibians. Genome 36:762–773
Martins C, Galetti PM (1999) Chromosomal localization of 5S rDNA genes in Leporinus fish (Anostomidae, Characiformes). Chromosome Res 7:363–367
Martins C, Wasko AP (2004) Organization and evolution of 5S ribosomal DNA in the fish genome. In: Williams CR (ed) Focus on genome research. Nova Science Publishers, Hauppauge, NY, USA, pp 289–318
Meyer JL, Shultz ET (1985) Migrating haemulid fishes as a source of nutrients and organic matter on coral reefs. Limnol Oceanogr 30:146–156
Nelson JS (1994) Fishes of the world, 3rd edn. John Wiley and Sons, Inc., New York
Nirchio M, Fenocchio AS, Swarça AC, Pérez JE, Granado A, Estrada A, Ron E (2003) Cytogenetic characterization of hybrids offspring between Colossoma macropomum (Cuvier, 1818) and Piaractus brachypomus (Cuvier, 1817) from Caicara del Orinoco, Venezuela. Caryologia 56(4):405–411
Nirchio M, Fenocchio AS, Swarça AC, Pérez JE (2004) Karyology of the toadfish Porichthys plectrodon (Jordan and Gilbert, 1882) (Batrachoididae) from Margarita Island, Venezuela. Mar Biol 146:161–165
Ohno S (1974) Protochordata, Cyclostomata and Pisces. In: John B (ed) Animal cytogenesis, vol 4, Chordata 1. Börntraeger, Berlin, pp 1–92
Pinkel D, Straume T, Gray JW (1986) Cytogenetic analysis using quantitative, high-sensitivity, fluorescence hybridization. PNAS 83:2934–2938
Randall JE (1996) Caribbean reef fishes, 3rd edn. T.H.F. Publications, Neptune City, NJ, USA
Reagan JD, Sigel MM, Lee HW, Llamass KA, Beasley AR (1968) Chromosomal alterations in marine fish cells in vitro. Can J Genet Cytol 10:448–453
Ron E, Nirchio M (2005) Caracterización citogenética de Haemulon flavolineatum Desmarest 1823 (Pisces: Haemulidae) de la Isla de Margarita, Venezuela. Bol Inst Oceanogr Univ Oriente 44(1):35–40
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, New York
Sola L, Cataudella S, Capanna E (1981) New developments in vertebrate cytotaxonomy. III. Karyology of bony fishes: a review. Genetica 54:285–328
Sumner AT (1972) A simple technique for demonstrating centromeric heterochromatin. Exp Cell Res 75:304–306
Suzuki H, Sakurai S, Matsuda Y (1996) Rat 5S rDNA spacer sequences and chromosomal assignment of the genes to the extreme terminal region of chromosome 19. Cytogenet Cell Genet 72:1–4
Vitturi R, Catalano E, Lo Conte MR, Alessi AM, Amico FP, Colombera D (1991) Intra-populational and intra-individual mosaicisms of Uranoscopus scaber L. (Perciformes, Uranoscopidae). Heredity 67:325–330
Acknowledgments
Funds supporting this study were provided by Consejo de Investigación of Universidad de Oriente, Venezuela. Support to C. Oliveira, I.A. Ferreira and C. Martins was granted by Fundação de Amparo à Pesquisa do Estado de São Paulo (FAPESP) Brazil and Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq) Brazil.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Nirchio, M., Gaviria, J.I., Oliveira, C. et al. Cytogenetic analysis of three species of the genus Haemulon (Teleostei: Haemulinae) from Margarita Island, Venezuela. Genetica 131, 135–140 (2007). https://doi.org/10.1007/s10709-006-9123-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10709-006-9123-4