Abstract
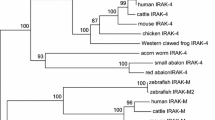
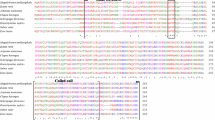
The ADAMs (a disintegrin and metalloproteinase) play regulatory roles in cell adhesion, migration and proteolysis. To explore the origin and evolution of ADAMs, this study identified the homologs of adam10 and adam17 in Lampetra morii and Lampetra japonica. Sequence analysis revealed that they share the same genomic structures with their counterparts in jawed vertebrates. The putative proteins possess conserved motifs, including a furin cut site (RXXR) for precursor processing, an enzyme catalytic motif (HEXGEHXXGXXH) for hydrolysis, and a Ca2+-binding motif (CGNXXXEXGEXCD) for stabilizing protein structure. In addition, a substrate recognition domain is present at the membrane-proximal region of lamprey ADAM17. The cytoplasmic region of lamprey ADAM10 contains a potential threonine phosphorylation site which has been shown to be activated by protein kinase C (PKC) in mammals. Both the adam10 and adam17 genes were constitutively expressed in the brain, kidney, and gills and were differentially regulated in the primary blood leukocytes by lipopolysaccharide (LPS) and pokeweed mitogen (PWM). Adam10 was induced by LPS but not PWM; conversely, adam17 was induced by PWM but not LPS. Taken together, our results suggest that the activation pathways and functions of ADAM10 and ADAM17 are conserved in agnathans.








Similar content being viewed by others
Data availability
The datasets generated during and/or analyzed during the current study are available from the corresponding authors on reasonable request.
Code availability
Not applicable.
Abbreviations
- BCR:
-
B-cell receptor
- CDS:
-
Complete coding sequence
- EGF:
-
Epidermal growth factor
- ErbB:
-
Epidermal growth factor receptor
- ERKs:
-
Extracellular regulated protein kinases
- GSP:
-
Gene-specific primer
- LPS:
-
Lipopolysaccharide
- MHC:
-
Major histocompatibility complex
- MPD:
-
Membrane-proximal domain
- NICD:
-
Notch intracellular domain
- PAMP:
-
Pathogen-associated molecular pattern
- PWM:
-
Pokeweed mitogen
- RACE:
-
Rapid amplification of cDNA end
- TACE:
-
TNFα-converting enzyme
- TCR:
-
T-cell receptor
- TM:
-
Transmembrane
- TNFα:
-
Tumor necrosis factor alpha
- UTR:
-
Untranslated region
- VLR:
-
Variable lymphocyte receptor
References
Anisimova M, Gil M, Dufayard J-F, Dessimoz C, Gascuel O (2011) Survey of branch support methods demonstrates accuracy, power, and robustness of fast likelihood-based approximation schemes. Syst Biol 60:685–699
Black RA, Rauch CT, Kozlosky CJ, Peschon JJ, Slack JL, Wolfson MF, Castner BJ, Stocking KL, Reddy P, Srinivasan S (1997) A metalloproteinase disintegrin that releases tumour-necrosis factor-α from cells. Nature 385:729–733
Bode W, Gomis-Rüth F-X, Stöckler W (1993) Astacins, serralysins, snake venom and matrix metalloproteinases exhibit identical zinc-binding environments (HEXXHXXGXXH and Met-turn) and topologies and should be grouped into a common family, the ‘metzincins.’ FEBS Lett 331:134–140
Boehm T, Hirano M, Holland SJ, Das S, Schorpp M, Cooper MD (2018) Evolution of alternative adaptive immune systems in vertebrates. Annu Rev Immunol 36:19–42
Cerretti D (1999) Characterization of the tumour necrosis factor α-converting enzyme, TACE/ADAM17. Biochem Soc Trans 27:219–223
Dazzo E, Leonardi E, Belluzzi E, Malacrida S, Vitiello L, Greggio E, Tosatto SC, Nobile C (2016) Secretion-positive LGI1 mutations linked to lateral temporal epilepsy impair binding to ADAM22 and ADAM23 receptors. PLoS Genet 12:e1006376
Dreymueller D, Pruessmeyer J, Groth E, Ludwig A (2012) The role of ADAM-mediated shedding in vascular biology. Eur J Cell Biol 91:472–485
Düsterhöft S, Michalek M, Kordowski F, Oldefest M, Sommer A, Röseler J, Reiss K, Grötzinger J, Lorenzen I (2015) Extracellular juxtamembrane segment of ADAM17 interacts with membranes and is essential for its shedding activity. Biochemistry 54:5791–5801
Evans JP, Schultz RM, Kopf GS (1995) Mouse sperm-egg plasma membrane interactions: analysis of roles of egg integrins and the mouse sperm homologue of PH-30 (fertilin) beta. J Cell Sci 108:3267–3278
Gascuel O (1997) BIONJ: an improved version of the NJ algorithm based on a simple model of sequence data. Mol Biol Evol 14:685–695
Guindon S, Dufayard J-F, Lefort V, Anisimova M, Hordijk W, Gascuel O (2010) New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst Biol 59:307–321
Han BW, Herrin BR, Cooper MD, Wilson IA (2008) Antigen recognition by variable lymphocyte receptors. Science 321:1834–1837
Heng TS, Painter MW, Elpek K, Lukacs-Kornek V, Mauermann N, Turley SJ, Koller D, Kim FS, Wagers AJ, Asinovski N (2008) The Immunological Genome Project: networks of gene expression in immune cells. Nat Immunol 9:1091–1094
Hirano M, Guo P, McCurley N, Schorpp M, Das S, Boehm T, Cooper MD (2013) Evolutionary implications of a third lymphocyte lineage in lampreys. Nature 501:435–438
Iwagaki A, Porro M, Pollack M (2000) Influence of synthetic antiendotoxin peptides on lipopolysaccharide (LPS) recognition and LPS-induced proinflammatory cytokine responses by cells expressing membrane-bound CD14. Infect Immun 68:1655–1663
Kasamatsu J, Oshiumi H, Matsumoto M, Kasahara M, Seya T (2010) Phylogenetic and expression analysis of lamprey toll-like receptors. Dev Comp Immunol 34:855–865
Keightley RG, Cooper MD, Lawton AR (1976) The T cell dependence of B cell differentiation induced by pokeweed mitogen. J Immunol 117:1538–1544
Kenny PA (2007) TACE: a new target in epidermal growth factor receptor dependent tumors. Differentiation 75:800–808
Lambrecht BN, Vanderkerken M, Hammad H (2018) The emerging role of ADAM metalloproteinases in immunity. Nat Rev Immunol 18:745–758
Li X, Fan H (2004) Loss of ectodomain shedding due to mutations in the metalloprotease and cysteine-rich/disintegrin domains of the tumor necrosis factor-α converting enzyme (TACE). J Biol Chem 279:27365–27375
Li Y, Xiao T, Zou J (2021) Fish TNF and TNF receptors. Sci China Life Sci 64:196–220
Lu X-J, Chen Q, Yang G-J, Chen J (2015) The TNFα converting enzyme (TACE) from ayu (Plecoglossus altivelis) exhibits TNFα shedding activity. Mol Immunol 63:497–504
Magee CN, Riella LV (2016) Notch and its ligands in alloimmunity and rejection. Curr Opin Organ Transplant 21:15–21
Moss ML, Minond D (2017) Recent advances in ADAM17 research: a promising target for cancer and inflammation. Mediat Inflamm 2017:9673537
Moss ML, Bomar M, Liu Q, Sage H, Dempsey P, Lenhart PM, Gillispie PA, Stoeck A, Wildeboer D, Bartsch JW (2007) The ADAM10 prodomain is a specific inhibitor of ADAM10 proteolytic activity and inhibits cellular shedding events. J Biol Chem 282:35712–35721
Pruessmeyer J, Ludwig A (2009) The good, the bad and the ugly substrates for ADAM10 and ADAM17 in brain pathology, inflammation and cancer. Semin Cell Dev Biol 20:164–174
Reiss K, Saftig P (2009) The “a disintegrin and metalloprotease”(ADAM) family of sheddases: physiological and cellular functions. Semin Cell Dev Biol 20:126–137
Saftig P, Lichtenthaler SF (2015) The alpha secretase ADAM10: a metalloprotease with multiple functions in the brain. Prog Neurobiol 135:1–20
Saftig P, Reiss K (2011) The “A Disintegrin And Metalloproteases” ADAM10 and ADAM17: novel drug targets with therapeutic potential? Eur J Cell Biol 90:527–535
Scheller J, Chalaris A, Garbers C, Rose-John S (2011) ADAM17: a molecular switch to control inflammation and tissue regeneration. Trends Immunol 32:380–387
Seals DF, Courtneidge SA (2003) The ADAMs family of metalloproteases: multidomain proteins with multiple functions. Genes Dev 17:7–30
Sepulcre MP, Alcaraz-Pérez F, López-Muñoz A, Roca FJ, Meseguer J, Cayuela ML, Mulero V (2009) Evolution of lipopolysaccharide (LPS) recognition and signaling: fish TLR4 does not recognize LPS and negatively regulates NF-κB activation. J Immunol 182:1836–1845
Soond SM, Everson B, Riches DW, Murphy G (2005) ERK-mediated phosphorylation of Thr735 in TNFα-converting enzyme and its potential role in TACE protein trafficking. J Cell Sci 118:2371–2380
Souza J, Lisboa A, Santos T, Andrade M, Neves V, Teles-Souza J, Jesus H, Bezerra T, Falcão V, Oliveira R (2020) The evolution of ADAM gene family in eukaryotes. Genomics 112:3108–3116
Subramanian B, Gao S, Lercher MJ, Hu S, Chen W-H (2019) Evolview v3: a webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res 47:W270–W275
Sun Z, Qin Y, Liu D, Wang B, Jia Z, Wang J, Gao Q, Zou J, Pang Y (2021) The evolution and functional characterization of CXC chemokines and receptors in lamprey. Dev Comp Immunol 116:103905
Takeda S (2016) ADAM and ADAMTS family proteins and snake venom metalloproteinases: a structural overview. Toxins 8:155
van Tetering G, van Diest P, Verlaan I, van der Wall E, Kopan R, Vooijs M (2009) Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J Biol Chem 284:31018–31027
Wetzel S, Seipold L, Saftig P (2017) The metalloproteinase ADAM10: a useful therapeutic target? Biochim Biophys Acta (BBA)-Mol Cell Res 186:20–20
Yang X, Si-Wei Z, Qing-Wei L (2016) Lamprey: a model for vertebrate evolutionary research. Zool Res 37:263
Funding
This work is funded by the National Key R&D Program of China (Grant No. 2018YFD0900302) and the National Natural Science Foundation of China (Grant Nos. 32030112 and U21A20268).
Author information
Authors and Affiliations
Contributions
Kaizheng Wu, Jing Xu: investigation, methodology, data curation, and writing—original draft. Zhao Jia, Junya Wang, Zixuan Wang, Jianhua Feng, Xiaozhen Zhu, Qin Liu, Bangjie Wang, Mingjie Li: investigation and methodology. Yue Pang: supervision, writing—review and editing. Jun Zou: conceptualization, funding acquisition, project administration, supervision, writing—review and editing.
Corresponding authors
Ethics declarations
Ethics approval
Experimental procedures were approved by the Animal Welfare and Research Ethics Committee of the Liaoning Normal University and the Institute of Dalian Medical University (Permit number: SYXK2004-0029).
Consent to participate
Not applicable.
Consent for publication
All authors have approved the manuscript for publication.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, K., Xu, J., Jia, Z. et al. Phylogeny and expression of ADAM10 and ADAM17 homologs in lamprey. Fish Physiol Biochem 49, 321–334 (2023). https://doi.org/10.1007/s10695-023-01184-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-023-01184-7