Abstract
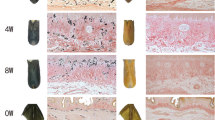
The commercial value of red tilapia is hampered by variations in skin color during overwintering. In this study, three types of skin of red tilapia, including the skin remained pink color during and after overwintering (P), the skin changed from pink color to black color during overwintering and remained black color after overwintering (P-B), and the skin changed from pink color to black color during overwintering but recovered to pink color when the temperature rose after overwintering (P-B-P), were used to analyze their molecular mechanisms of color variation. The transcriptome results revealed that the P, P-B, and P-B-P libraries had 43, 42, and 43 million clean reads, respectively. The top 10 abundance mRNAs and specific mRNAs (specificity measure SPM > 0.9) were screened. After comparing intergroup gene expression levels, there were 2528, 1924, and 1939 differentially expressed genes (DEGs) between P-B-P and P-B, P-B-P and P, and P-B and P, respectively. Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses of color-related mRNAs showed that a number of DEGs, including tyrp1, tyr, pmel, mitf, mc1r, asip, tat, hpdb, and foxd3, might play a potential role in pigmentation. Additionally, the co-expression patterns of genes were detected within the pigment-related pathways by the PPI network from P-B vs. P group. Furthermore, DEGs from the apoptosis and autophagy pathways, such as baxα, beclin1, and atg7, might be involved in the fading of red tilapia melanocytes. The findings will aid in understanding the molecular mechanism underlying skin color variation in red tilapia during and after overwintering as well as lay a foundation for future research aimed at improving red tilapia skin color characteristics.





Similar content being viewed by others
Availability of data and material
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Change history
27 June 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10695-022-01092-2
References
Andrews S (2014) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 20 June 2021
Bian FF, Yang XF, Ou ZJ et al (2019) Morphological characteristics and comparative transcriptome analysis of three different phenotypes of Pristella maxillaris. Front Genet 10:698
Braasch I, Liedtke D, Volff JN et al (2010) Pigmentary function and evolution of tyrp1 gene duplicates in fish. Pigment Cell Melanoma Res 22(6):839–850
Ceinos RM, Guillot R, Kelsh RN et al (2015) Pigment patterns in adult fish result from superimposition of two largely independent pigmentation mechanisms. Pigment Cell Melanoma Res 28(2):196–209
Cheli Y, Ohanna M, Ballotti R, Bertolotto C (2010) Fifteen-year quest for microphthalmia-associated transcription factor target genes. Pigment Cell Melanoma Res 23(1):27–40
Chen G, Shi Y, Liu M et al (2018a) circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis 9(2):175
Chen YX, Chen YS, Shi CM et al (2018b) Soapnuke: a mapreduce acceleration supported software for integrated quality control and preprocessing of high-throughput sequencing data. Oxford Open 7(1):gix120
Dong ZJ, Luo MK, Wang LM et al (2020) MicroRNA-206 regulation of skin pigmentation in koi carp (Cyprinus carpio L.). Front Genet 11:47
Fernandez PJ, Bagnara JT (1991) Effect of background color and low temperature on skin color and circulating α-MSH in two species of leopard frog. Gen Comp Endocrinol 83(1):132–141
Franz M, Lopes CT, Huck G et al (2016) Cytoscape.Js a graph theory library for visualisation and analysis. Bioinformatics 32(2):309–311
Fujimura N, Taketo MM, Mori M et al (2009) Spatial and temporal regulation of Wnt/β-catenin signaling is essential for development of the retinal pigment epithelium. Dev Biol 334(1):31–45
Gouveia L, Rema P (2005) Effect of microalgal biomass concentration and temperature on ornamental goldfish (Carassius auratus) skin pigmentation. Aquac Nutr 11(1):19–23
Gross JB, Borowsky R, Tabin CJ (2009) A novel role for mc1r in the parallel evolution of depigmentation in independent populations of the cavefish Astyanax Mexicanus. PLoS Genet 5(1):1000326
Guillot R, Ceinos RM, Cal R et al (2012) Transient ectopic overexpression of agouti-signalling protein 1 (asip1) induces pigment anomalies in flatfish. PLoS One 7(12):e48526
Gupta MV, Acosta BO (2004) A review of global tilapia farming practices. Aquacult Asia Pac 9(1):7–12
Gutierrez GB, Wiener P, Williams JL (2007) Genetic effects on coat color in cattle: dilution of eumelanin and phaeomelanin pigments in an F2-backcross Charolais ×Holstein population. BMC Genet 8(1):56
Henning F, Jones JC, Franchini P et al (2013) Transcriptomics of morphological color change in polychromatic Midas cichlids. BMC Genomics 14(1):171–171
Higdon CW, Mitra RD, Johnson SL et al (2013) Gene expression analysis of zebrafish melanocytes, iridophores, and retinal pigmented epithelium reveals indicators of biological function and developmental origin. PLoS One 8(7):e67801
Hubbard JK, Uy JA, Hauber ME et al (2010) Vertebrate pigmentation: from underlying genes to adaptive function. Trends Genet 26:231–239
Jiang BJ, Fu JJ, Dong ZJ et al (2019) Maternal ancestry analyses of red tilapia strains based on D-loop sequences of seven tilapia populations. PeerJ 7:e7007
Jiang YL, Zhang SH, Xu J et al (2014) Comparative transcriptome analysis reveals the genetic basis of skin color variation in common carp. PLoS One 9(9):e108200
José MCR, Tatjana H, Helgi BS et al (2005) Gene structure of the goldfish agouti-signaling protein: a putative role in the dorsal-ventral pigment pattern of fish. Endocrinology 146(3):1597–1610
Kats LB, Van DRG (1986) Background color-matching in the spring peeper. Hyla Crucifer 1:109–115
Kim D, Langmead B, Salzberg SL (2015) Hisat: a fast spliced aligner with low memory requirements. Nat Methods 12(4):357–360
Komatsu M, Waguri S, Ueno T et al (2005) Impairment of starvation-induced and constitutive autophagy in atg7 deficient mice. J Cell Biol 169(3):425–434
Krauss J, Geiger-Rudolph S, Koch I et al (2015) A dominant mutation in tyrp1a leads to melanophore death in zebrafish. Pigm Cell Melanoma Res 27(5):827–830
Li XJ, Li SF, Feng JH et al (2003) Preliminary study on salinity tolerance of Israel red tilapia. J Shanghai Fish Univ 12(3):205–208
Li XM, Song YN, Xiao GB et al (2015) Gene expression variations of red-white skin coloration in common carp (Cyprinus carpio). Int J Mol Sci 16:21310–21329
Liao Y, Gordon K, Smyth et al (2014) Featurecounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30(7):923–930
Lister JA, Close J, Raible DW (2001) Duplicate mitf genes in zebrafish: complementary expression and conservation of melanogenic potential. Dev Biol 237(2):333–344
Liu CS, Liu LY, Zhang Y et al (2020) Molecular mechanism of aqp3 in regulating differentiation and apoptosis of lung cancer stem cells through Wnt/GSK-3β/β-Catenin pathway. Jbuon 25(4):1714–1720
Luo MK, Lu GQ, Yin HR et al (2021) Fish pigmentation and coloration: molecular mechanisms and aquaculture perspectives. Rev Aquac 00:1–18
Minvielle F, Bed’Hom B, Coville JL et al (2010) The “silver” Japanese quail and the mitf gene: causal mutation, associated traits and homology with the “blue” chicken plumage. BMC Genet 11(1):1–7
Oltvai ZN, Milliman CL, Korsmeyer SJ (1993) Bcl-2 heterodimerizes in vivo with a conserved homolog, bax, that accelerates programed cell death. Cell 74(4):609–619
Pan JB, Hu SC, Wang H et al (2012) PaGeFinder: quantitative identification of spatiotemporal pattern genes. Bioinformatics 28(11):1544–1545
Parmar MB, Venkatachalam AB, Wright JM (2012) The evolutionary relationship of the transcriptionally active fabp11a (intronless) and fabp11b genes of medaka with fabp11 genes of other teleost fishes. FEBS J 279(13):2310–2321
Pavlidis M, Karkana M, Fanouraki E et al (2008) Environmental control of skin color in the red porgy, Pagrus Pagrus. Aquac Res 39(8):837–849
Pietrobono S, Anichini G, Sala C et al (2020) ST3GAL1 is a target of the SOX2-GLI1 transcriptional complex and promotes melanoma metastasis through AXL. Nat Commun 11(1):5865
Pradeep PJ, Srijaya TC, Hassan A et al (2014) Optimal conditions for cold-shock induction of triploidy in red tilapia. Aquacult Int 22(3):1163–1174
Richardson J, Lundegaard PR, Reynolds NL, Dorin JR, Porteous DJ, Jackson IJ, Patton EE (2008) Mc1r pathway regulation of zebrafish melanosome dispersion. Zebrafish 5(4):289–295
Robinson MD, Mccarthy DJ, Smyth GK (2010) Edger: a bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26(1):139–140
Sherbrooke WC, Frost SK (1989) Integumental chromatophores of a color-change, thermoregulating lizard, Phrynosoma modestum (Iguanidae; reptilia). Am Mus Novit 2943(2):1–14
Simon DJ, Peles D, Wakamatsu K et al (2009) Current challenges in understanding melanogenesis: bridging chemistry, biological control, morphology and function. Pigment Cell Melanoma Res 22(5):563–579
Skarnes WC, Rosen B, West AP et al (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474(7351):337–342
Solano F, Martinez-Esparza M, Jimenez-Cervantes C, Hill SP, Lozano JA, Garcia-Borron JC (2000) New insights on the structure of the mouse silver locus and on the function of the silver protein. Pigment Cell Res 13(8):118–124
Swiercz JM, Worzfeld T, Offermanns S (2008) ErbB-2 and met reciprocally regulate cellular signaling via plexin-B1. J Biol Chem 283(4):1893–1901
Szklarczyk D, Franceschini A, Wyder S et al (2015) STRING v10: protein-protein networks, integrated over the tree of life. Nucleic Acids Res 43:D447–D452
Takacs-Vellai K, Vellai T, Puoti A et al (2005) Inactivation of the autophagy gene bec-1 triggers apoptotic cell death in C. elegans. Curr Biol 15(16):1513–1517
Tezuka A, Yamamoto H, Yokoyama J et al (2011) The mc1r gene in the guppy (Poecilia reticulata): genotypic and phenotypic polymorphisms. BMC Res Notes 4(1):31
Thomas AJ, Erickson CA (2009) Foxd3 regulates the lineage switch between neural crest-derived glial cells and pigment cells by repressing mitf through a non-canonical mechanism. Development 136(11):1849–1858
Voisey J, Box NF, Daal AV (2001) A polymorphism study of the human agouti gene and its association with mc1r. Pigment Cell Res 14(4):264–267
Wang LG, Wang SQ, Li W (2012) RSeQC: quality control of RNA-seq experiments. Bioinformatics 28(16):2184–2185
Wang LM, Song FB, Zhu WB et al (2018) Effects of temperature on body color of Malaysian red tilapia during overwintering period. J Fish China 42(1):72–79
Wang XY, Yang J, Yao Y et al (2020) Aqp3 facilitates proliferation and adipogenic differentiation of porcine intramuscular adipocytes. Genes 11(4):453
Wardani WW, Alimuddin A, Junior MZ et al (2020) Growth performance, robustness against stress, serum insulin, igf-1 and glut4 gene expression of red tilapia (Oreochromis sp.) fed diet containing graded levels of creatine. Aquac Nutr 27(1):274–286
Wu X, Zhao JD, Ruan YY et al (2018) Sialyltransferase st3gal1 promotes cell migration, invasion, and TGF-β1-induced EMT and confers paclitaxel resistance in ovarian cancer. Cell Death Dis 9(11):1102
Xie C, Mao XZ, Huang JJ et al (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucleic Acids Res 39(suppl_2):W316-22
Yu GC, Wang LG, Han YY et al (2012) Clusterprofiler: an r package for comparing biological themes among gene clusters. OMICS 16(5):284–287
Zhang Y, Wang L, Zhang S et al (2008) HmgA, transcriptionally activated by hpda, influences the biosynthesis of actinorhodin in Streptomyces Coelicolor. FEMS Microbiol Lett 280(2):219–225
Zhang YP, Wang ZD, Guo YS et al (2015) Morphological characters and transcriptome profiles associated with black skin and red skin in crimson snapper (Lutjanus erythropterus). Int J Mol Sci 16(11):26991–27004
Zhang YQ, Liu JH, Peng LY et al (2017) Comparative transcriptome analysis of molecular mechanism underlying gray-to-red body color formation in red crucian carp (Carassius auratus, red var.). Fish Physiol Biochem 43(6):1387–1398
Zhu WB, Wang LM, Dong ZJ et al (2016) Comparative transcriptome analysis identifies candidate genes related to skin color differentiation in red tilapia. Sci Rep 6(1):31347
Funding
This study was supported by the National Natural Science Foundation-Youth Fund Project (31802290).
Author information
Authors and Affiliations
Contributions
Zaijie Dong conceived the study; Bingjie Jiang performed the experiments and wrote the paper; Wenbin Zhu provided the experimental materials; Lanmei Wang provided the funding for the experiment; Jianjun Fu and Wei Liu provided technical assistance in the experiments; Mingkun Luo revised the manuscript; Zaijie Dong reviewed the manuscript; all authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
The sampling scheme and experimental protocols were subject to approval by the Bioethical Committee of Freshwater Fisheries Research Center (FFRC) of the Chinese Academy of Fishery Sciences (CAFS) (BC 2013863, 9/2013). The methods of samples handled and experimental procedures carried out in accordance with the guidelines for the care and use of animals for scientific purposes issued by the Ministry of Science and Technology, Beijing, China (No.398, 2006).
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Jiang, B., Wang, L., Luo, M. et al. Transcriptome analysis of skin color variation during and after overwintering of Malaysian red tilapia. Fish Physiol Biochem 48, 669–682 (2022). https://doi.org/10.1007/s10695-022-01073-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-022-01073-5