Abstract
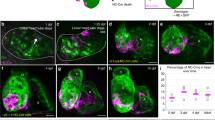
As a tightly controlled biological process, cardiogenesis requires the specification and migration of a suite of cell types to form a particular three-dimensional configuration of the heart. Many genetic factors are involved in the formation and maturation of the heart, and any genetic mutations may result in severe cardiac failures. The neuron navigator (NAV) family consists of three vertebrate homologs (NAV1, NAV2, and NAV3) of the neural guidance molecule uncoordinated-53 (UNC-53) in Caenorhabditis elegans. Although they are recognized as neural regulators, their expressions are also detected in many organs, including the heart, kidney, and liver. However, the functions of NAVs, regardless of neural guidance, remain largely unexplored. In our study, we found that nav3 gene was expressed in the cardiac region of zebrafish embryos from 24 to 48 h post-fertilization (hpf) by means of in situ hybridization (ISH) assay. A CRISPR/Cas9-based genome editing method was utilized to delete the nav3 gene in zebrafish and loss of function of Nav3 resulted in a severe deficiency in its cardiac morphology and structure. The similar phenotypic defects of the knockout mutants could recur by nav3 morpholino injection and be rescued by nav3 mRNA injection. Dual-color fluorescence imaging of ventricle and atrium markers further confirmed the disruption of the heart development in nav3-deleted mutants. Although the heart rate was not affected by the deletion of nav3, the heartbeat intensity was decreased in the mutants. All these findings indicate that Nav3 was required for cardiogenesis in developing zebrafish embryos.




Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
Code availability
Not applicable.
Abbreviations
- CH:
-
Calponin homology
- dpf:
-
Days post-fertilization
- HI:
-
Heartbeat intensity
- hpf:
-
Hours post-fertilization
- ISH:
-
In situ hybridization
- NAV:
-
Neuron navigator
- SH3:
-
Src homology 3
- UNC53:
-
Uncoordianted-53
- WISH:
-
Whole-mount in situ hybridization
- WT:
-
Wild-type
References
Bakkers J (2011) Zebrafish as a model to study cardiac development and human cardiac disease. Cardiovasc Res 91:279–288
Berdougo E, Coleman H, Lee DH, Stainier DYR, Yelon D (2003) Mutation of weak atrium/atrial myosin heavy chain disrupts atrial function and influences ventricular morphogenesis in zebrafish. Development 130:6121–6129
Buckingham M, Meilhac S, Zaffran S (2005) Building the mammalian heart from two sources of myocardial cells. Nat Rev Genet 6:826–835
Carlsson E, Krohn K, Ovaska K, Lindberg P, Häyry V, Maliniemi P, Lintulahti A, Korja M, Kivisaari R, Hussein S et al (2013) Neuron navigator 3 alterations in nervous system tumors associate with tumor malignancy grade and prognosis. Genes Chromosomes Cancer 52:191–201
Cohen-Dvashi H, Ben-Chetrit N, Russell R, Carvalho S, Lauriola M, Nisani S, Mancini M, Nataraj N, Kedmi M, Roth L et al (2015) Navigator-3, a modulator of cell migration, may act as a suppressor of breast cancer progression. EMBO Mol Med 7:299–314
Gore AV, Monzo K, Cha YR, Pan W, Weinstein BM (2012) Vascular development in the zebrafish. Cold Spring Harb Perspect Med 2:1–21
Grassini, D.R., Lagendijk, A.K., De Angelis, J.E., Da Silva, J., Jeanes, A., Zettler, N., Bower, N.I., Hogan, B.M., and Smith, K.A. (2018). Nppa and nppb act redundantly during zebrafish cardiac development to confine AVC marker expression and reduce cardiac jelly volume. Dev. Camb. 145.
Haworth KE, Kotecha S, Mohun TJ, Latinkic BV (2008) GATA4 and GATA5 are essential for heart and liver development in Xenopus embryos. BMC Dev Biol 8:1–16
Holtzinger A, Evans T (2007) Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev Biol 312:613–622
Jiao K, Kulessa H, Tompkins K, Zhou Y, Batts L, Baldwin HS, Hogan BLM (2003) An essential role of Bmp4 in the atrioventricular septation of the mouse heart. Genes Dev 17:2362–2367
Karenko L, Hahtola S, Päivinen S, Karhu R, Syrjä S, Kähkönen M, Nedoszytko B, Kytölä S, Zhou Y, Blazevic V et al (2005) Primary cutaneous T-cell lymphomas show a deletion or translocation affecting NAV3, the human UNC-53 homologue. Cancer Res 65:8101–8110
Keegan BR, Meyer D, Yelon D (2004) Organization of cardiac chamber progenitors in the zebrafish blastula. Development 131:3081–3091
Klein C, Mikutta J, Krueger J, Scholz K, Brinkmann J, Liu D, Veerkamp J, Siegel D, Abdelilah-Seyfried S, le Noble F (2011) Neuron navigator 3a regulates liver organogenesis during zebrafish embryogenesis. Development 138:1935–1945
Krueger J, Liu D, Scholz K, Zimmer A, Shi Y, Klein C, Siekmann A, Schulte-Merker S, Cudmore M, Ahmed A et al (2011) Flt1 acts as a negative regulator of tip cell formation and branching morphogenesis in the zebrafish embryo. Development 138:2111–2120
Liu J, Stainier DYR (2012) Zebrafish in the study of early cardiac development. Circ Res 110:870–874
Lv F, Zhu C, Yan X, Wang X, Liu D (2017) Generation of a mef2aa:EGFP transgenic zebrafish line that expresses EGFP in muscle cells. Fish Physiol Biochem 43:287–294
Maes T, Barceló A, Buesa C (2002) Neuron navigator: a human gene family with homology to UNC-53, a cell guidance gene from Caenorhabditis elegans. Genomics 80:21–30
Mittal N, Yoon SH, Enomoto H, Hiroshi M, Shimizu A, Kawakami A, Fujita M, Watanabe H, Fukuda K, Makino S (2019) Versican is crucial for the initiation of cardiovascular lumen development in medaka (Oryzias latipes). Sci Rep 9:1–17
Reiter JF, Alexander J, Rodaway A, Yelon D, Patient R, Holder N, Stainier DYR (1999) Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev 13:2983–2995
Rueden CT, Schindelin J, Hiner MC, DeZonia BE, Walter AE, Arena ET, Eliceiri KW (2017) Image J2: ImageJ for the next generation of scientific image data. BMC Bioinformatics 18:529
Schindler YL, Garske KM, Wang J, Firulli BA, Firulli AB, Poss KD, Yelon D (2014) Hand2 elevates cardiomyocyte production during zebrafish heart development and regeneration. Dev Camb 141:3112–3122
Schmidt KL, Marcus-Gueret N, Adeleye A, Webber J, Baillie D, Stringham EG (2009) The cell migration molecule UNC-53/NAV2 is linked to the ARP2/3 complex by ABI-1. Development 136:563–574
Stringham E, Pujol N, Vandekerckhove J, Bogaert T (2002) Unc-53 controls longitudinal migration in C. elegans. Development 129:3367–3379
Targoff KL, Colombo S, George V, Schell T, Kim SH, Solnica-Krezel L, Yelon D (2013) Nkx genes are essential for maintenance of ventricular identity. Dev Camb 140:4203–4213
Yelon D, Horne SA, Stainier DYR (1999) Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev Biol 214:23–37
Zhang R, Xu X (2009) Transient and transgenic analysis of the zebrafish ventricular myosin heavy chain (vmhc) promoter: an inhibitory mechanism of ventricle-specific gene expression. Dev Dyn 238:1564–1573
Funding
The research conducted in this manuscript was funded by the National Natural Science Foundation of China (82000458) to CC and start-up funding for Doctoral Research of Nantong Science and Technology College (NTKY-Dr2017001) to FL. It is also funded by Nantong Science and Technology Program (JC2021088), Open Program of Key Laboratory of Cultivation and High-value Utilization of Marine Organisms in Fujian Province (2019fjsccq08), Natural Science Foundation of the Jiangsu Higher Education Institutions of China (20KJB180008), and the Science Foundation of Nantong City (JC2020023).
Author information
Authors and Affiliations
Contributions
CC and DL designed and conceptualized the project. FL, XG, PQ, and XL performed the experiment. FL and CC analyzed the data. CC wrote the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval
All animal-related experiments were carried out following the NIH Guidelines for the care and use of laboratory animals (http://oacu.od.nih.gov/regs/index.htm), and animal protocols were ethically approved by the Administration Committee of Experimental Animals of Nantong University, Jiangsu Province, China (Approval ID: 20180608-Z001). Transgenic zebrafish lines provided by others were approved by the owners with written informed consent.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Disclaimer
The funding sources provided financial support for the experiments described, but had no role in the design of the study or collection, analysis, and interpretation of data or in writing the manuscript.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Feng Lv, Xiaojuan Ge and Peipei Qian are contributed equally to this work.
Supplementary Information
Below is the link to the electronic supplementary material.
10695_2022_1049_MOESM1_ESM.png
Supplementary Figure S1. Expressions of cardiac-related genes in WT and nav3-/- mutants at 24 hpf. The expression of the corresponding gene is outlined with a white dashed line. The digits at the lower left position of each panel indicate the number of fishes with typical phenotype in total observed ones. (PNG 3028 KB)
Rights and permissions
About this article
Cite this article
Lv, F., Ge, X., Qian, P. et al. Neuron navigator 3 (NAV3) is required for heart development in zebrafish. Fish Physiol Biochem 48, 173–183 (2022). https://doi.org/10.1007/s10695-022-01049-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10695-022-01049-5