Abstract
Missense variants of DNA mismatch repair (MMR) genes pose a problem in clinical genetics as long as they cannot unambiguously be assigned as the cause of Lynch syndrome (LS). To study such variants of uncertain clinical significance, we have developed a functional assay based on direct measurement of MMR activity in mouse embryonic stem cells expressing mutant protein from the endogenous alleles. We have applied this protocol to a specific truncation mutant of MSH2 that removes 60 C-terminal amino acids and has been found in suspected LS families. We show that the stability of the MSH2/MSH6 heterodimer is severely perturbed, causing attenuated MMR in in vitro assays and cancer predisposition in mice. This mutation can therefore unambiguously be considered as deleterious and causative for LS.




Similar content being viewed by others
References
Lynch HT, Lynch J (2000) Lynch syndrome: genetics, natural history, genetic counseling, and prevention. J Clin Oncol 18:19S–31S
Ionov Y, Peinado MA, Malkhosyan S, Shibata D, Perucho M (1993) Ubiquitous somatic mutations in simple repeated sequences reveal a new mechanism for colonic carcinogenesis. Nature 363:558–561
Nash GM, Gimbel M, Shia J, Culliford AT, Nathanson DR, Ndubuisi M, Yamaguchi Y, Zeng ZS, Barany F, Paty PB (2003) Automated, multiplex assay for high-frequency microsatellite instability in colorectal cancer. J Clin Oncol 21:3105–3112
Karran P (2001) Mechanisms of tolerance to DNA damaging therapeutic drugs. Carcinogenesis 22:1931–1937
Stojic L, Mojas N, Cejka P, di Pietro M, Ferrari S, Marra G, Jiricny J (2003) Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev 18:1331–1344
Rayssiguier C, Thaler DS, Radman M (1989) The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature 342:396–401
De Wind N, Dekker M, Berns A, Radman M, te Riele H (1995) Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell 82:321–330
Gradia S, Subramanian D, Wilson T, Acharya S, Makhov A, Griffith J, Fishel R (1999) hMSH2-hMSH6 forms a hydrolysis-independent sliding clamp on mismatched DNA. Mol Cell 3:255–261
Umar A, Risinger JI, Glaab WE, Tindall KR, Barrett JC, Kunkel TA (1998) Functional overlap in mismatch repair by human MSH3 and MSH6. Genetics 148:1637–1646
Dekker M, de Vries S, Aarts M, Dekker R, Brouwers C, Wiebenga O, de Wind N, Cantelli E, Tonelli R, te Riele H (2011) Transient suppression of MLH1 allows effective single-nucleotide substitution by single-stranded oligonucleotides. Mutat Res 715:52–60
Wielders EA, Dekker RJ, Holt I, Morris GE, te Riele H (2011) Characterization of MSH2 variants by endogenous gene modification in mouse embryonic stem cells. Hum Mutat 32:389–396
Mangold E, Pagenstecher C, Friedl W, Mathiak M, Buettner R, Engel C, Loeffler M, Holinski-Feder E, Müller-Koch Y, Keller G, Schackert HK, Krüger S, Goecke T, Moeslein G, Kloor M, Gebert J, Kunstmann E, Schulmann K, Rüschoff J, Propping P (2005) Spectrum and frequencies of mutations in MSH2 and MLH1 identified in 1,721 German families suspected of hereditary nonpolyposis colorectal cancer. Int J Cancer 116:692–702
Krüger S, Plaschke J, Jeske B, Görgens H, Pistorius SR, Bier A, Kreuz FR, Theissig F, Aust DE, Saeger HD, Schackert HK (2003) Identification of six novel MSH2 and MLH1 germline mutations in HNPCC. Hum Mutat 21:445–446
Wang Q, Lasset C, Desseigne F, Saurin JC, Maugard C, Navarro C, Ruano E, Descos L, Trillet-Lenoir V, Bosset JF, Puisieux A (1999) Prevalence of germline mutations of hMLH1, hMSH2, hPMS1, hPMS2, and hMSH6 genes in 75 French kindreds with nonpolyposis colorectal cancer. Hum Genet 105:79–85
Papp J, Kovacs ME, Olah E (2007) Germline MLH1 and MSH2 mutational spectrum including frequent large genomic aberrations in Hungarian hereditary non-polyposis colorectal cancer families: implications for genetic testing. World J Gastroenterol 13:2727–2732
Miyaki M, Konishi M, Muraoka M, Kikuchi-Yanoshita R, Tanaka K, Iwama T, Mori T, Koike M, Ushio K, Chiba M et al (1995) Germ line mutations of hMSH2 and hMLH1 genes in Japanese families with hereditary nonpolyposis colorectal cancer (HNPCC): usefulness of DNA analysis for screening and diagnosis of HNPCC patients. J Mol Med 73:515–520
Yuan Y, Han HJ, Zheng S, Park JG (1998) Germline mutations of hMLH1 and hMSH2 genes in patients with suspected hereditary nonpolyposis colorectal cancer and sporadic early-onset colorectal cancer. Dis Colon Rectum 41:434–440
Millar AL, Pal T, Madlensky L, Sherman C, Temple L, Mitri A, Cheng H, Marcus V, Gallinger S, Redston M, Bapat B, Narod S (1999) Mismatch repair gene defects contribute to the genetic basis of double primary cancers of the colorectum and endometrium. Hum Mol Genet 8:823–829
Terdiman JP, Gum JR Jr, Conrad PG, Miller GA, Weinberg V, Crawley SC, Levin TR, Reeves C, Schmitt A, Hepburn M, Sleisenger MH, Kim YS (2001) Efficient detection of hereditary nonpolyposis colorectal cancer gene carriers by screening for tumor microsatellite instability before germline genetic testing. Gastroenterology 120:21–30
Durno C, Aronson M, Bapat B, Cohen Z, Gallinger S (2005) Family history and molecular features of children, adolescents, and young adults with colorectal carcinoma. Gut 54:1146–1150
Bianchi F, Galizia E, Bracci R, Belvederesi L, Catalani R, Loretelli C, Giorgetti G, Ferretti C, Bearzi I, Porfiri E, Cellerino R (2007) Effectiveness of the CRCAPRO program in identifying patients suspected for HNPCC. Clin Genet 71:158–164
Baudhuin LM, Mai M, French AJ, Kruckeberg KE, Swanson RL, Winters JL, Courteau LK, Thibodeau SN (2005) Analysis of hMLH1 and hMSH2 gene dosage alterations in hereditary nonpolyposis colorectal cancer patients by novel methods. J Mol Diagn 7:226–235
Percesepe A, Borghi F, Menigatti M, Losi L, Foroni M, Di Gregorio C, Rossi G, Pedroni M, Sala E, Vaccina F, Roncucci L, Benatti P, Viel A, Genuardi M, Marra G, Kristo P, Peltomäki P, Ponz de Leon M (2001) Molecular screening for hereditary nonpolyposis colorectal cancer: a prospective, population-based study. J Clin Oncol 19:3944–3950
Ponz de Leon M, Benatti P, Borghi F, Pedroni M, Scarselli A, Di Gregorio C, Losi L, Viel A, Genuardi M, Abbati G, Rossi G, Menigatti M, Lamberti I, Ponti G, Roncucci L (2004) Aetiology of colorectal cancer and relevance of monogenic inheritance. Gut 53:115–122
Hampel H, Frankel WL, Martin E, Arnold M, Khanduja K, Kuebler P, Nakagawa H, Sotamaa K, Prior TW, Westman J, Panescu J, Fix D, Lockman J, Comeras I, de la Chapelle A (2005) Screening for the Lynch syndrome (hereditary nonpolyposis colorectal cancer). N Engl J Med 352:1851–1860
Wagner A, Barrows A, Wijnen JT, van der Klift H, Franken PF, Verkuijlen P, Nakagawa H, Geugien M, Jaghmohan-Changur S, Breukel C, Meijers-Heijboer H, Morreau H, van Puijenbroek M, Burn J, Coronel S, Kinarski Y, Okimoto R, Watson P, Lynch JF, de la Chapelle A, Lynch HT, Fodde R (2003) Molecular analysis of hereditary nonpolyposis colorectal cancer in the United States: high mutation detection rate among clinically selected families and characterization of an American founder genomic deletion of the MSH2 gene. Am J Hum Genet 72:1088–1100
Swensen J, Lewis CM, Cannon-Albright LA (1997) Identification of a one-base germline deletion (codon 888 del C) and an intron splice acceptor site polymorphism in hMSH2. Hum Mutat 10:80–81
Warren JJ, Pohlhaus TJ, Changela A, Iyer RR, Modrich PL, Beese LS (2007) Structure of the human MutSalpha DNA lesion recognition complex. Mol Cell 26:579–592
Gupta S, Gellert M, Yang W (2012) Mechanism of mismatch recognition revealed by human MutSβ bound to unpaired DNA loops. Nat Struct Mol Biol 19:72–78
Lamers MH, Perrakis A, Enzlin JH, Winterwerp HH, de Wind N, Sixma TK (2000) The crystal structure of DNA mismatch repair protein MutS binding to a G x T mismatch. Nature 407:711–717
Calmann MA, Nowosielska A, Marinus MG (2005) The MutS C terminus is essential for mismatch repair activity in vivo. J Bacteriol 187:6577–6579
Mendillo ML, Putnam CD, Kolodner RD (2007) Escherichia coli MutS tetramerization domain structure reveals that stable dimers but not tetramers are essential for DNA mismatch repair in vivo. J Biol Chem 282:16345–16354
Dekker M, Brouwers C, Aarts M, van der Torre J, de Vries S, van de Vrugt H, te Riele H (2006) Effective oligonucleotide-mediated gene disruption in ES cells lacking the mismatch repair protein MSH3. Gene Ther 13:686–694
De Wind N, Dekker M, Claij N, Jansen L, van Klink Y, Radman M, Riggins G, Van der Valk M, Van ‘t Wout K, Te Riele H (1999) HNPCC-like cancer predisposition in mice through simultaneous loss of Msh3 and Msh6 mismatch-repair protein functions. Nat Genet 23:359–362
Claij N, Te Riele H (2002) Methylation tolerance in mismatch repair proficient cells with low MSH2 level. Oncogene 21:2873–2879
Acknowledgements
We thank dr. Allan Bradley for providing the Msh2 −/− ; Msh3 −/− ESC line, Roelof Pruntel for help with sequencing and microsatellite analysis, Marieke Aarts, Marleen Dekker and Hellen Houlleberghs for discussions and helpful comments on the manuscript, and members of the NKI animal facility for technical support. This work was financially supported by the Dutch Cancer Society (Grant NKI 2009-4477).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Fig. S1
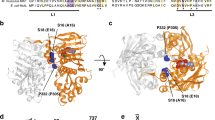
Three-dimensional structure of MSH2/MSH3 dimer. The image is derived from Gupta et al. [29] and shows MSH2 on the right and MSH3 on the left. The three cyan and two grey helices at the bottom constitute dimerization domains at the C-termini of MSH2 and MSH3, respectively, which contribute to dimer stabilization by hydrophobic interactions and salt bridges. Mutations listed in Table S1 disconnect the most right cyan helix from the lower brown helix in MSH2 that are linked by a disordered (non-visible) loop (PNG 514 kb)
Rights and permissions
About this article
Cite this article
Wielders, E., Delzenne-Goette, E., Dekker, R. et al. Truncation of the MSH2 C-terminal 60 amino acids disrupts effective DNA mismatch repair and is causative for Lynch syndrome. Familial Cancer 16, 221–229 (2017). https://doi.org/10.1007/s10689-016-9945-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-016-9945-x