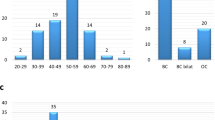
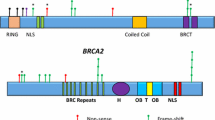
Abstract
Germline mutations in BRCA1 and BRCA2 cause hereditary breast and ovarian cancer. Molecular screening of these two genes in patients with a family history of breast or ovarian cancer has revealed pathogenic variants as well as genetic variants of unknown significance (VUS). These VUS may cause a challenge in the genetic counseling process regarding clinical management of the patient and the family. We investigated 32 variants previously detected in 33 samples from patients with a family history of breast or ovarian cancer. cDNA was analyzed for alternative transcripts and selected missense variants located in the BRCT domains of BRCA1 were assessed for their trans-activation ability. Although an extensive cDNA analysis was done, only three of the 32 variants appeared to affect the splice-process (BRCA1 c.213-5T>A, BRCA1 c.5434C>G and BRCA2 c.68-7T>A). In addition, two variants located in the BRCT domains of BRCA1 (c.5075A>C p.Asp1692Ala and c.5513T>G p.Val1838Gly) were shown to abolish the BRCT domain trans-activation ability, whereas BRCA1 c.5125G>A p.Gly1709Arg exhibited equal trans-activation capability as the WT domain. These functional studies may offer further insights into the pathogenicity of certain identified variants; however, this assay is only applicable for a subset of missense variants.


Similar content being viewed by others
References
Safran M, Dalah I, Alexander J, et al (2010) GeneCards Version 3: the human gene integrator. Database (Oxford) 2010: baq020 DOI 10.1093/database/baq020
Roy R, Chun J, Powell SN (2012) BRCA1 and BRCA2: different roles in a common pathway of genome protection. Nat Rev Cancer 12(1):68–78. doi:10.1038/nrc3181
Oliver AW, Swift S, Lord CJ, Ashworth A, Pearl LH (2009) Structural basis for recruitment of BRCA2 by PALB2. EMBO Rep 10(9):990–996. doi:10.1038/embor.2009.126
Kast K, Rhiem K, Wappenschmidt B et al (2016) Prevalence of BRCA1/2 germline mutations in 21 401 families with breast and ovarian cancer. J Med Genet 53(7):465–471. doi:10.1136/jmedgenet-2015-103672
Frank TS, Deffenbaugh AM, Reid JE, et al (2002) Clinical characteristics of individuals with germline mutations in BRCA1 and BRCA2: analysis of 10,000 individuals. J Clin Oncol 20(6):1480–1490
Barnes DR, Antoniou AC (2012) Unravelling modifiers of breast and ovarian cancer risk for BRCA1 and BRCA2 mutation carriers: update on genetic modifiers. J Intern Med 271(4):331–343. doi:10.1111/j.1365-2796.2011.02502.x
Gaildrat P, Krieger S, Thery JC et al (2010) The BRCA1 c.5434C → G (p. Pro1812Ala) variant induces a deleterious exon 23 skipping by affecting exonic splicing regulatory elements. J Med Genet 47(6):398–403. doi:10.1136/jmg.2009.074047
Friedman LS, Ostermeyer EA, Szabo CI et al (1994) Confirmation of BRCA1 by analysis of germline mutations linked to breast and ovarian cancer in ten families. Nat Genet 8(4):399–404. doi:10.1038/ng1294-399
Hoffman JD, Hallam SE, Venne VL, Lyon E, Ward K (1998) Implications of a novel cryptic splice site in the BRCA1 gene. Am J Med Genet 80(2):140–144
Fetzer S, Tworek HA, Piver MS, Dicioccio RA (1998) An alternative splice site junction in exon 1a of the BRCA1 gene. Cancer Genet Cytogenet 105(1):90–92
Colombo M, Blok MJ, Whiley P et al (2014) Comprehensive annotation of splice junctions supports pervasive alternative splicing at the BRCA1 locus: a report from the ENIGMA consortium. Hum Mol Genet 23(14):3666–3680. doi:10.1093/hmg/ddu075
Jakubowska A, Gorski B, Byrski T et al (2001) Detection of germline mutations in the BRCA1 gene by RNA-based sequencing. Hum Mutat 18(2):149–156. doi:10.1002/humu.1164
Fackenthal JD, Yoshimatsu T, Zhang B et al (2016) Naturally occurring BRCA2 alternative mRNA splicing events in clinically relevant samples. J Med Genet. doi:10.1136/jmedgenet-2015-103570
Narod SA, Foulkes WD (2004) BRCA1 and BRCA2: 1994 and beyond. Nat Rev Cancer 4(9):665–676. doi:10.1038/nrc1431
Carvalho MA, Marsillac SM, Karchin R et al (2007) Determination of cancer risk associated with germ line BRCA1 missense variants by functional analysis. Cancer Res 67(4):1494–1501. doi:10.1158/0008-5472.CAN-06-3297
den Dunnen JT, Antonarakis SE (2000) Mutation nomenclature extensions and suggestions to describe complex mutations: a discussion. Hum Mutat 15(1):7–12. doi:10.1002/(SICI)1098-1004(200001)15:1<7:AID-HUMU4>3.0.CO;2-N
Untergasser A, Cutcutache I, Koressaar T et al (2012) Primer3—new capabilities and interfaces. Nucl Acids Res 40(15):e115. doi:10.1093/nar/gks596
Koressaar T, Remm M (2007) Enhancements and modifications of primer design program Primer3. Bioinformatics 23(10):1289–1291. doi:10.1093/bioinformatics/btm091
Sequencher® version 5.3 sequence analysis software. Gene Codes Corporation, Ann Arbor, MI USA. http://www.genecodes.com
Lindor NM, Guidugli L, Wang X et al (2012) A review of a multifactorial probability-based model for classification of BRCA1 and BRCA2 variants of uncertain significance (VUS). Hum Mutat 33(1):8–21. doi:10.1002/humu.21627
Cherbal F, Salhi N, Bakour R, Adane S, Boualga K, Maillet P (2012) BRCA1 and BRCA2 unclassified variants and missense polymorphisms in Algerian breast/ovarian cancer families. Dis Markers 32(6):343–353. doi:10.3233/DMA-2012-0893
Spurdle AB, Whiley PJ, Thompson B et al (2012) BRCA1 R1699Q variant displaying ambiguous functional abrogation confers intermediate breast and ovarian cancer risk. J Med Genet 49(8):525–532. doi:10.1136/jmedgenet-2012-101037
Bouwman P, van der Gulden H, van der Heijden I et al (2013) A high-throughput functional complementation assay for classification of BRCA1 missense variants. Cancer Discov 3(10):1142–1155. doi:10.1158/2159-8290.CD-13-0094
Lovelock PK, Spurdle AB, Mok MT et al (2007) Identification of BRCA1 missense substitutions that confer partial functional activity: Potential moderate risk variants? Breast Cancer Res 9(6):R82. doi:10.1186/bcr1826
Kelemen O, Convertini P, Zhang Z et al (2013) Function of alternative splicing. Gene 514(1):1–30. doi:10.1016/j.gene.2012.07.083
Easton DF, Deffenbaugh AM, Pruss D et al (2007) A systematic genetic assessment of 1433 sequence variants of unknown clinical significance in the BRCA1 and BRCA2 breast cancer-predisposition genes. Am J Hum Genet 81(5):873–883. doi:10.1086/521032
Cooper TA, Mattox W (1997) The regulation of splice-site selection, and its role in human disease. Am J Hum Genet 61(2):259–266. doi:10.1086/514856
Hoya M, Soukarieh O, Lopez-Perolio I et al (2016) Combined genetic and splicing analysis of BRCA1 c.[594-2A>C; 641A>G] highlights the relevance of naturally occurring in-frame transcripts for developing disease gene variant classification algorithms. Hum Mol Genet. doi:10.1093/hmg/ddw094
Palacios IM (2013) Nonsense-mediated mRNA decay: from mechanistic insights to impacts on human health. Brief Funct Genom 12(1):25–36. doi:10.1093/bfgp/els051
Buisson M, Anczukow O, Zetoune AB, Ware MD, Mazoyer S (2006) The 185delAG mutation (c.68_69delAG) in the BRCA1 gene triggers translation reinitiation at a downstream AUG codon. Hum Mutat 27(10):1024–1029. doi:10.1002/humu.20384
Shakya R, Reid LJ, Reczek CR et al (2011) BRCA1 tumor suppression depends on BRCT phosphoprotein binding, but not its E3 ligase activity. Science 334(6055):525–528. doi:10.1126/science.1209909
Vreeswijk MP, Kraan JN, van der Klift HM et al (2009) Intronic variants in BRCA1 and BRCA2 that affect RNA splicing can be reliably selected by splice-site prediction programs. Hum Mutat 30(1):107–114. doi:10.1002/humu.20811
Sanz DJ, Acedo A, Infante M et al (2010) A high proportion of DNA variants of BRCA1 and BRCA2 is associated with aberrant splicing in breast/ovarian cancer patients. Clin Cancer Res 16(6):1957–1967. doi:10.1158/1078-0432.CCR-09-2564
Xia B, Sheng Q, Nakanishi K et al (2006) Control of BRCA2 cellular and clinical functions by a nuclear partner, PALB2. Mol Cell 22(6):719–729. doi:10.1016/j.molcel.2006.05.022
Santos C, Peixoto A, Rocha P et al (2014) Pathogenicity evaluation of BRCA1 and BRCA2 unclassified variants identified in Portuguese breast/ovarian cancer families. J Mol Diagn 16(3):324–334. doi:10.1016/j.jmoldx.2014.01.005
Houdayer C, Caux-Moncoutier V, Krieger S et al (2012) Guidelines for splicing analysis in molecular diagnosis derived from a set of 327 combined in silico/in vitro studies on BRCA1 and BRCA2 variants. Hum Mutat 33(8):1228–1238. doi:10.1002/humu.22101
Lee MS, Green R, Marsillac SM et al (2010) Comprehensive analysis of missense variations in the BRCT domain of BRCA1 by structural and functional assays. Cancer Res 70(12):4880–4890. doi:10.1158/0008-5472.CAN-09-4563
Jhuraney A, Velkova A, Johnson RC et al (2015) BRCA1 Circos: a visualisation resource for functional analysis of missense variants. J Med Genet 52(4):224–230. doi:10.1136/jmedgenet-2014-102766
Lek M, Karczewski K, Minikel E, et al (2015) Analysis of protein-coding genetic variation in 60,706 humans. Biorxiv. doi:10.1101/030338
Simard J, Dumont M, Moisan AM et al (2007) Evaluation of BRCA1 and BRCA2 mutation prevalence, risk prediction models and a multistep testing approach in French-Canadian families with high risk of breast and ovarian cancer. J Med Genet 44(2):107–121. doi:10.1136/jmg.2006.044388
Castera L, Krieger S, Rousselin A et al (2014) Next-generation sequencing for the diagnosis of hereditary breast and ovarian cancer using genomic capture targeting multiple candidate genes. Eur J Hum Genet 22(11):1305–1313. doi:10.1038/ejhg.2014.16
Millot GA, Berger A, Lejour V et al (2011) Assessment of human Nter and Cter BRCA1 mutations using growth and localization assays in yeast. Hum Mutat 32(12):1470–1480. doi:10.1002/humu.21608
Scottish/Northern Irish BBC (2003) BRCA1 and BRCA2 mutations in Scotland and Northern Ireland. Br J Cancer 88(8):1256–1262. doi:10.1038/sj.bjc.6600840
Bonnet C, Krieger S, Vezain M et al (2008) Screening BRCA1 and BRCA2 unclassified variants for splicing mutations using reverse transcription PCR on patient RNA and an ex vivo assay based on a splicing reporter minigene. J Med Genet 45(7):438–446. doi:10.1136/jmg.2007.056895
Schoumacher F, Glaus A, Mueller H, Eppenberger U, Bolliger B, Senn HJ (2001) BRCA1/2 mutations in Swiss patients with familial or early-onset breast and ovarian cancer. Swiss Med Wkly 131(15–16):223–226
Barker DF, Almeida ER, Casey G et al (1996) BRCA1 R841 W: a strong candidate for a common mutation with moderate phenotype. Genet Epidemiol 13(6):595–604. doi:10.1002/(SICI)1098-2272(1996)13:6<595:AID-GEPI5>3.0.CO;2-#
Durocher F, Shattuck-Eidens D, McClure M et al (1996) Comparison of BRCA1 polymorphisms, rare sequence variants and/or missense mutations in unaffected and breast/ovarian cancer populations. Hum Mol Genet 5(6):835–842
Goldgar DE, Easton DF, Deffenbaugh AM et al (2004) Integrated evaluation of DNA sequence variants of unknown clinical significance: application to BRCA1 and BRCA2. Am J Hum Genet 75(4):535–544. doi:10.1086/424388
Panguluri RC, Brody LC, Modali R et al (1999) BRCA1 mutations in African Americans. Hum Genet 105(1–2):28–31
van Orsouw NJ, Dhanda RK, Elhaji Y et al (1999) A highly accurate, low cost test for BRCA1 mutations. J Med Genet 36(10):747–753
Vallon-Christersson J, Cayanan C, Haraldsson K et al (2001) Functional analysis of BRCA1 C-terminal missense mutations identified in breast and ovarian cancer families. Hum Mol Genet 10(4):353–360
Scott CL, Jenkins MA, Southey MC et al (2003) Average age-specific cumulative risk of breast cancer according to type and site of germline mutations in BRCA1 and BRCA2 estimated from multiple-case breast cancer families attending Australian family cancer clinics. Hum Genet 112(5–6):542–551. doi:10.1007/s00439-003-0908-6
Laraqui A, Uhrhammer N, Lahlou-Amine I et al (2013) Mutation screening of the BRCA1 gene in early onset and familial breast/ovarian cancer in Moroccan population. Int J Med Sci 10(1):60–67. doi:10.7150/ijms.5014
Chenevix-Trench G, Healey S, Lakhani S et al (2006) Genetic and histopathologic evaluation of BRCA1 and BRCA2 DNA sequence variants of unknown clinical significance. Cancer Res 66(4):2019–2027. doi:10.1158/0008-5472.CAN-05-3546
Martinez-Ferrandis JI, Vega A, Chirivella I et al (2003) Mutational analysis of BRCA1 and BRCA2 in Mediterranean Spanish women with early-onset breast cancer: identification of three novel pathogenic mutations. Hum Mutat 22(5):417–418. doi:10.1002/humu.9188
Hilton JL, Geisler JP, Rathe JA, Hattermann-Zogg MA, DeYoung B, Buller RE (2002) Inactivation of BRCA1 and BRCA2 in ovarian cancer. J Natl Cancer Inst 94(18):1396–1406
Thery JC, Krieger S, Gaildrat P et al (2011) Contribution of bioinformatics predictions and functional splicing assays to the interpretation of unclassified variants of the BRCA genes. Eur J Hum Genet 19(10):1052–1058. doi:10.1038/ejhg.2011.100
Guidugli L, Carreira A, Caputo SM et al (2014) Functional assays for analysis of variants of uncertain significance in BRCA2. Hum Mutat 35(2):151–164. doi:10.1002/humu.22478
Stegel V, Krajc M, Zgajnar J et al (2011) The occurrence of germline BRCA1 and BRCA2 sequence alterations in Slovenian population. BMC Med Genet 12:9. doi:10.1186/1471-2350-12-9
Wagner TM, Hirtenlehner K, Shen P et al (1999) Global sequence diversity of BRCA2: analysis of 71 breast cancer families and 95 control individuals of worldwide populations. Hum Mol Genet 8(3):413–423
Kanchi KL, Johnson KJ, Lu C et al (2014) Integrated analysis of germline and somatic variants in ovarian cancer. Nat Commun 5:3156. doi:10.1038/ncomms4156
Karchin R, Agarwal M, Sali A, Couch F, Beattie MS (2008) Classifying Variants of Undetermined Significance in BRCA2 with protein likelihood ratios. Cancer Inform 6:203–216
Guidugli L, Pankratz VS, Singh N et al (2013) A classification model for BRCA2 DNA binding domain missense variants based on homology-directed repair activity. Cancer Res 73(1):265–275. doi:10.1158/0008-5472.CAN-12-2081
Llort G, Munoz CY, Tuser MP et al (2002) Low frequency of recurrent BRCA1 and BRCA2 mutations in Spain. Hum Mutat 19(3):307. doi:10.1002/humu.9014
Menendez M, Castellsague J, Mirete M et al (2012) Assessing the RNA effect of 26 DNA variants in the BRCA1 and BRCA2 genes. Breast Cancer Res Treat 132(3):979–992. doi:10.1007/s10549-011-1661-5
Acknowledgments
We thank Alvaro N. A. Monteiro for kindly providing us with the BRCT containing plasmids necessary for the trans-activation assay. We also thank “Helse Nord” for providing the necessary funding for this study (Grant # SFP1161-14).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Informed consent
All participants gave written informed consent for diagnostic testing. The project was submitted to the appropriate regional ethics committee, however, since the samples were tested diagnostically the regional ethical committee waved the need for ethical approval based on the Norwegian regional health organization law § 2 and § 9 and the Norwegian research ethical law § 4.
Rights and permissions
About this article
Cite this article
Jarhelle, E., Riise Stensland, H.M.F., Mæhle, L. et al. Characterization of BRCA1 and BRCA2 variants found in a Norwegian breast or ovarian cancer cohort. Familial Cancer 16, 1–16 (2017). https://doi.org/10.1007/s10689-016-9916-2
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-016-9916-2