Abstract
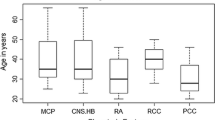
von Hippel–Lindau (VHL) disease is a hereditary tumor syndrome caused by mutations in the VHL tumor suppressor gene. In a family with VHL, we identified a novel missense mutation (N78D), which affects a fully conserved residue in the VHL protein. Interestingly, several other missense mutations reported at same codon in the VHL protein that might be associated with a low risk of renal cell carcinoma (RCC) but not pheochromocytoma appear to be associated with a VHL type 1 phenotype. At the moment, RCC is present in none of the affected mutation carriers in the family described here. In contrast to other missense changes at codon 78, the change in our VHL family is predicted to have a mild effect on VHL function, which apparently is insufficient to cause predisposition to RCC. Our findings suggest that the risk of RCC in VHL is attributable to the severity of the amino acid substitution at this particular codon in the VHL protein.



Similar content being viewed by others
References
Richards FM, Maher ER, Latif F, Phipps ME, Tory K, Lush M, Crossey PA, Oostra B, Gustavson KH, Green J, Turner G, Yates JRW, Linehan WM, Affara NA, Lerman M, Zbar B, Ferguson-Smith MA (1993) Detailed genetic mapping of the von Hippel–Lindau disease tumour suppressor gene. J Med Genet 30:104–107
Glenn GM, Daniel LN, Choyke P, Linehan WM, Oldfield E, Gorin MB, Hosoe S, Latif F, Weiss G, Walther M, Lerman MI, Zbar B (1991) Von Hippel–Lindau (VHL) disease: distinct phenotypes suggest more than one mutant allele at the VHL locus. Hum Genet 87:207–210
Maher ER, Yates JRW, Ferguson-Smith MA (1990) Statistical analysis of the two stage mutation model in von Hippel-Lindau disease, and in sporadic cerebellar haemangioblastoma and renal cell carcinoma. J Med Genet 27:311–314
Lonser RR, Glenn GM, Walther M, Chew EY, Libutti SK, Linehan WM, Oldfield EH (2003) von Hippel–Lindau disease. Lancet 361:2059–2067
Neumann HPH, Wiestler OD (1991) Clustering of features of von Hippel–Lindau syndrome: evidence for a complex genetic locus. Lancet 337:1052–1054
Brauch H, Kishida T, Glavac D, Chen F, Pausch F, Hofler H, Latif F, Lerman MI, Zbar B, Neumann HPH (1995) Von Hippel–Lindau (VHL) disease with pheochromocytoma in the Black Forest region of Germany: evidence for a founder effect. Hum Genet 95:551–556
Hoffman MA, Ohh M, Yang H, Klco JM, Ivan M, Kaelin WG Jr (2001) von Hippel–Lindau protein mutants linked to type 2C VHL disease preserve the ability to downregulate HIF. Hum Mol Genet 10:1019–1027
Nordstrom-O’Brien M, van der Luijt RB, van Rooijen E, van den Ouweland AM, Majoor-Krakauer DF, Lolkema MP, van Brussel A, Voest EE, Giles RH (2010) Genetic analysis of von Hippel–Lindau disease. Hum Mutat 31:521–537
Clifford SC, Cockman ME, Smallwood AC, Mole DR, Woodward ER, Maxwell PH, Ratcliffe PJ, Maher ER (2001) Contrasting effects on HIF-1alpha regulation by disease-causing pVHL mutations correlate with patterns of tumourigenesis in von Hippel–Lindau disease. Hum Mol Genet 10(10):1029–1038
Bangiyeva V, Rosenbloom A, Alexander AE, Isanova B, Popko T, Schoenfeld AR (2009) Differences in regulation of tight junctions and cell morphology between VHL mutations from disease subtypes. BMC Cancer 14(9):229
Crossey PA, Richards FM, Foster K, Green JS, Prowse A, Latif F, Lerman MI, Zbar B, Affara NA, Ferguson-Smith MA, Maher ER (1994) Identification of intragenic mutations in the von Hippel–Lindau disease tumour suppressor gene and correlation with disease phenotype. Hum Mol Genet 3(8):1303–1308
Chen F, Kishida T, Yao M, Hustad T, Glavac D, Dean M, Gnarra JR, Orcutt ML, Duh FM, Glenn G, Green J, Hsia YE, Lamiell J, Li H, Wei MH, Schmidt L, Tory K, Kuzman I, Stackhouse T, Latif F, Linehan WM, Lerman M, Zbar B (1995) Germline mutations in the von Hippel–Lindau disease tumor suppressor gene: correlations with phenotype. Hum Mutat 5:66–75
Zbar B, Kishida T, Chen F, Schmidt L, Maher ER, Richards FM, Crossey PA, Webster AR, Affara NA, Ferguson-Smith MA, Brauch H, Glavac D, Neumann HPH, Tisherman S, Mulvihill JJ, Gross D, Shuin T, Whaley J, Seizinger B, Kley N, Olschwang S, Boisson C, Richard S, Lips CHM, Linehan MW, Lerman M (1996) Germline mutations in the Von Hippel–Lindau disease (VHL) gene in families from North America, Europe, and Japan. Hum Mutat 8:348–357
Neumann HP, Bender BU (1998) Genotype-phenotype correlations in von Hippel–Lindau disease. J Intern Med 243(6):541–545
Forman JR, Worth CL, Bickerton GR, Eisen TG, Blundell TL (2009) Structural bioinformatics mutation analysis reveals genotype-phenotype correlations in von Hippel–Lindau disease and suggests molecular mechanisms of tumorigenesis. Proteins 77(1):84–96
Stebbins CE, Kaelin WG Jr, Pavletich NP (1999) Structure of the VHL-ElonginC-ElonginB complex: implications for VHL tumor suppressor function. Science 284(5413):455–461
Liu J, Nussinov R (2008) Allosteric effects in the marginally stable von Hippel–Lindau tumor suppressor protein and allostery-based rescue mutant design. Proc Natl Acad Sci USA 105(3):901–906
Hes FJ, van der Luijt RB, Janssen ALW, Zewald RA, de Jong GJ, Lenders JW, Links TP, Luyten GPM, Sijmons RH, Eussen HJ, Halley DJJ, Lips CJM, Pearson PL, van den Ouweland AMW, Majoor-Krakauer DF (2007) Frequency of Von Hippel–Lindau germline mutations in classic and non-classic Von Hippel–Lindau disease identiWed by DNA sequencing, Southern blot analysis and multiplex ligation-dependent probe ampliWcation. Clin Genet 72:122–129
Ong KR, Woodward ER, Killick P, Lim C, Macdonald F, Maher ER (2007) Genotype-phenotype correlations in von Hippel–Lindau disease. Hum Mutat 28:143–149
Gossage L, Cartwright E, Eisen T, Bycroft MA (2010) Alterations in VHL as potential biomarkers in renal-cell carcinoma. Nat Rev Clin Oncol 7(5):277–288
McNeill A, Rattenberry E, Barber R, Killick P, MacDonald F, Maher ER (2009) Genotype-phenotype correlations in VHL exon deletions. Am J Med Genet 149A:2147–2151
Gomy I, Molfetta GA, de Andrade Barreto E, Ferreira CA, Zanette DL, Casali-da-Rocha JC, Silva WA Jr (2010) Clinical and molecular characterization of Brazilian families with von Hippel–Lindau disease: a need for delineating genotype-phenotype correlation. Fam Cancer 9(4):635–642
Tavtigian SV, Deffenbaugh AM, Yin L, Judkins T, Scholl T, Samollow PB, de Silva D, Zharkikh A, Thomas A (2006) Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J Med Genet 43(4):295–305
Kishida T, Stackhouse TM, Chen F, Lerman MI, Zbar B (1995) Cellular proteins that bind the von Hippel–Lindau disease gene product: mapping of binding domains and the effect of missense mutations. Cancer Res 55:4544–4548
Kanno H, Shuin T, Kondo K, Ito S, Hosaka M, Torigoe S, Fujii S, Tanaka Y, Yamamoto I, Kim I, Yao M (1996) Molecular genetic diagnosis of von Hippel–Lindau disease: analysis of five Japanese families. Jpn J Cancer Res 87:423–428
Clinical Research Group for VHL in Japan (1995) Germline mutations in the von Hippel–Lindau disease (VHL) gene in Japanese VHL. Hum Mol Genet 4:2233–2237
Bailly M, Bain C, Favrot MC, Ozturk M (1995) Somatic mutations of Von Hippel–Lindau (VHL) tumor-supressor gene in european kidney cancers. Int J Cancer 63:660–664
Maher ER, Webster AR, Richards FM, Green JS, Crossey PA, Payne SJ, Moore AT (1996) Phenotypic expression in von Hippel–Lindau disease: correlations with germline VHL gene mutations. J Med Genet 33:328–332
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
10689_2012_9586_MOESM1_ESM.doc
Supplementary Figure S1 Localization of the VHL gene mutation identified in our study. Sequence electropherograms showing c.232A >G transition resulting in substitution of Asn by Asp at codon 78 (N78D)
Rights and permissions
About this article
Cite this article
Cingoz, S., van der Luijt, R.B., Kurt, E. et al. A novel missense mutation (N78D) in a family with von Hippel–Lindau disease with central nervous system haemangioblastomas, pancreatic and renal cysts. Familial Cancer 12, 111–117 (2013). https://doi.org/10.1007/s10689-012-9586-7
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10689-012-9586-7