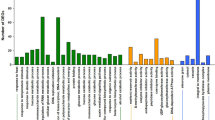
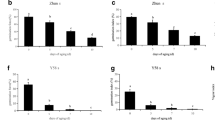
Abstract
Seed aging is a complex and irreversible process that occurs during seed development and storage. The quality of Allium mongolicum Regel seeds directly impacts its cultivation and production. However, the mechanism of aging seeds in A. mongolicum Regel is not well understood. Germination and physiological indicators were assessed, and RNA-Seq technique was performed on samples stored for 1, 4, and 8 years. In total, 5723, 7347 and 5293 differentially expressed genes (DEGs) were identified in 1 year old (S1) versus 4 year old (S2), S1 versus 8 year old (S4), and S2 versus S4, respectively. The number of DEGs increased as the storage time increased. The analysis revealed that the majority of the these DEGs were involved in a wide range of processes, such as protein synthesis, degradation and targeting; RNA processing and regulation; transport; DNA; signaling; stress; metabolism; amino acid metabolism; secondary metabolism; and hormone metabolism. The germination rate of A. mongolicum seeds remained consistently high, exceeding 80%, even after four years of storage. This observation indicates a potential correlation with the increased expression levels of aspartic proteases. However, as the storage time surpassed the seed’s lifespan, there was a notable decline in the expression levels of genes that encode aspartic proteases, cysteine proteases, and serine proteases. Conducting further research on these candidate genes could enhance our comprehension of the regulatory mechanisms involved in seed aging during storage for varying durations.








Similar content being viewed by others
References
Bentsink L, Alonso-Blanco C, Vreugdenhil D, Tesnier K, Groot SPC, Koornneef M (2000) Genetic analysis of seed-soluble oligosaccharides in relation to seed storability of Arabidopsis. Plant Physiol 124:1595–1604. https://doi.org/10.1104/pp.124.4.1595
Blauer JM, Knowles LO, Knowles NR (2013) Evidence that tuber respiration is the pacemaker of physiological aging in seed potatoes (Solanum tuberosum L.). J Plant Growth Regul 32:708–720. https://doi.org/10.1007/s00344-013-9338-4
Booth DT, Sowa S (2001) Respiration in dormant and non-dormant bitterbrush seeds. J Arid Environ 48:35–39. https://doi.org/10.1006/jare.2000.0737
Chen RZ (1986) Determination of the activity of acidic phosphatase in seeds. Seed 4:79–53
Chen H, Osuna D, Colville L, Lorenzo O, Graeber K, Kster H, Leubner-Metzger G, Kranner I (2013) Transcriptome-wide mapping of pea seed ageing reveals a pivotal role for genes related to oxidative stress and programmed cell Death. PLoS ONE 10:e78471. https://doi.org/10.1371/journal.pone.0078471
Chen HJ, Huang YH, Huang GJ, Huang SS, Chow TJ, Lin YH (2015) Sweet potato SPAP1 is a typical aspartic protease and participates in ethephon-mediated leaf senescence. J Plant Physiol 180:1–17. https://doi.org/10.1016/j.jplph.2015.03.009
Chen Y, Ding Z, Wu Y, Chen Q, Liu M, Yu H, Wang D, Zhang Y, Wang T (2020) Effects of Allium mongolicum Regel and its flavonoids on constipation. Biomolecules 10:14–28. https://doi.org/10.3390/biom10010014
Cheng WH, Chiang MH, Hwang SG, Lin PL (2009) Antagonism between abscisic acid and ethylene in Arabidopsis acts in parallel with the reciprocal regulation of their metabolism and signaling pathways. Plant Mol Biol 71:61–80. https://doi.org/10.1007/s11103-009-9509-7
Cruz de Carvalho MH, d’Arcy-Lameta A, Roy-Macauley H, Gareil M, El Maarouf H, Pham-Thi AT, Zuily-Fodil Y (2001) Aspartic protease in leaves of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata L. Walp): enzymatic activity, gene expression and relation to drought susceptibility. FEBS Lett 492:242–246. https://doi.org/10.1016/S0014-5793(01)02259-1
Cuniberti A, Tolley A, Riglos MVC, Giovachini R (2010) Influence of natural aging on the precipitation hardening of an AlMgSi alloy. Mat SciI Eng A 527:5307–5311. https://doi.org/10.1016/j.msea.2010.05.003
Deng QJ, Liu JH, Jin ZQ, Xu BY (2011) Changes of malate dehydrogenase and malic acid content during the ripening of banana fruit. Chin J Trop Agric 31:34–38
Fenollosa E, Jené L, Munné-Bosch S (2020) A rapid and sensitive method to assess seed longevity through accelerated aging in an invasive plant species. Plant Methods 16:64–74. https://doi.org/10.1186/s13007-020-00607-3
Filipenko EA, Kochetov AV, Kanayama Y, Malinovsky VI, Shumny VK (2013) PR-proteins with ribonuclease activity and plant resistance against pathogenic fungi. Russ J Genet Appl Res 3:474–480. https://doi.org/10.1134/S2079059713060026
Gho YS, Park SA, Kim SR, Chandran AKN, An G, Jung KH (2017) Comparative expression analysis of rice and Arabidopsis peroxiredoxin genes suggests conserved or diversified roles between the two species and leads to the identification of tandemly duplicated rice peroxiredoxin genes differentially expressed in seeds. Rice 10:30–43. https://doi.org/10.1186/s12284-017-0170-5
Govender V, Aveling TAS, Kritzinger Q (2008) The effect of traditional storage methods on germination and vigour of maize (Zea mays L.) from northern KwaZulu-Natal and southern Mozambique. S Afr J Bot 74:190–196. https://doi.org/10.1016/j.sajb.2007.10.006
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol 29:644–652. https://doi.org/10.1038/nbt.1883
Hang NT, Lin Q, Liu L, Liu X, Liu S, Wang W, Li L, He N, Liu Z, Jiang L, Wan J (2015) Mapping QTLs related to rice seed storability under natural and artificial aging storage conditions. Euphytica 203:673–681. https://doi.org/10.1007/s10681-014-1304-0
Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell DevBiol 19:141–172. https://doi.org/10.1146/annurev.cellbio.19.110701.154617
Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S (2002) RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nat 419:135–141. https://doi.org/10.1038/nature00991
Hu BN, Yang ZR, Wang P, Zhang FL, Chen GH, Li XJ, Zhao QY, Hao LZ (2018) Study on nutrient component content of Allium mongolicum Regel seed. J Anhui Agr Sci 13:5376–5377
Hu QJ, Chen MX, Song T, Cheng CL, Tian Y, Hu J, Zhang JH (2020) Spermidine enhanced the antioxidant capacity of rice seeds during seed aging. Plant Growth Regul 91:397–406. https://doi.org/10.1007/s10725-020-00613-4
Jenner HL, Winning BM, Millar AH, Tomlinson KL, Leaver CJ, Hill SA (2001) NAD malic enzyme and the control of carbohydrate metabolism in potato tubers. Plant Physiol 126:1139–1150. https://doi.org/10.1104/pp.126.3.1139
Kurek K, Plitta-Michalak B, Ratajczak E (2019) Reactive oxygen species as potential drivers of the seed aging process. Plants (basel) 8:174–186. https://doi.org/10.3390/plants8060174
Laloi M, McCarthy J, Morandi O, Gysler C, Bucheli P (2002) Molecular and biochemical characterisation of two aspartic proteinases TcAP1 and TcAP2 from Theobroma cacao seeds. Planta 215:754–762. https://doi.org/10.1007/s00425-002-0818-1
Landjeva S, Lohwasser U, Börner A (2010) Genetic mapping within the wheat D genome reveals QTL for germination, seed vigour and longevity, and early seedling growth. Euphytica 171:129–143. https://doi.org/10.1007/s10681-009-0016-3
Lee J, Welti R, Roth M, Schapaugh WT, Li J, Trick HN (2012) Enhanced seed viability and lipid compositional changes during natural ageing by suppressing phospholipase Dα in soybean seed. Plant Biotechnol J 10:164–173. https://doi.org/10.1111/j.1467-7652.2011.00650.x
Lennon JT, Jones SE (2011) Microbial seed banks: the ecological and evolutionary implications of dormancy. Nat Rev Microbiol 9:119–130. https://doi.org/10.1038/nrmicro2504
Liang Y, Liu XY, Zhang HW (2014) Cloning and expression analysis of the anthocyanin biosynthesis-related gene AcPAL1 in onion. Chin J Agric Biotechol 22:47–54
Linkies A, Müller K, Morris K, Turečková V, Wenk M, Cadman CSC, Corbineau F, Strnad M, Lynn JR, Finch-Savage WE, Leubner-Metzger G (2009) Ethylene interacts with abscisic acid to regulate endosperm rupture during germination: a comparative approach using Lepidium sativum and Arabidopsis thaliana. Plant Cell 21:3803–3822. https://doi.org/10.1105/tpc.109.070201
Liu X, Hu P, Huang M, Tang Y, Li Y, Li L, Hou X (2016) The NF-YC–RGL2 module integrates GA and ABA signalling to regulate seed germination in Arabidopsis. Nat Commun 7:12768–12781. https://doi.org/10.1038/ncomms12768
Liu H, Zhu YF, Liu X, Jiang Y, Deng SM, Ai X, Deng ZJ (2020a) Effect of artificially accelerated aging on the vigor of Metasequoia glyptostroboides seeds. J for Res 31:769–779. https://doi.org/10.1007/s11676-018-0840-1
Liu Y, He J, Yan Y, Liu A, Zhang HQ (2020b) Comparative transcriptomic analysis of two rice (Oryza sativa L.) male sterile line seed embryos under accelerated aging. Plant Mol Biol Rep 38:282–293. https://doi.org/10.1007/s11105-020-01198-y
Makale A, Nene W, Kapinga F, Ndibanya K (2020) Effects of cashew seeds storage duration on germination and performance of seedlings in Tanzania. Am J Plant Sci 11:1784–1796. https://doi.org/10.4236/ajps.2020.1111128
McDonald MB (1999) Seed deterioration: physiology repair and assessment. Seed Sci Technol 27:177–237. https://doi.org/10.1016/j.jbiotec.2008.07.318116_Y-027
Michel H (1999) Cytochrome c oxidase: catalytic cycle and mechanisms of proton pumping-a discussion. Biochemistry 38:15129–15140. https://doi.org/10.1021/bi9910934
Minic Z (2008) Physiological roles of plant glycoside hydrolases. Planta 227:723–740
Parkhey S, Naithani KS (2012) ROS production and lipid catabolism in desiccating Shorea robusta seeds during aging. Plant Physiol Biochem 57:261–267. https://doi.org/10.1016/j.plaphy.2012.06.008
Pereira MD, Dias D, Borges E, Filho S, Dias LAS, Soriano P (2013) Physiological quality of physic nut (Jatropha curcas L.) seeds during storage. J Seed Sci 35:21–27. https://doi.org/10.1590/S2317-15372013000100003
Rajjou L, Lovigny Y, Groot SP, Belghazi M, Job C, Job D (2008) Proteome-wide characterization of seed aging in Arabidopsis: a comparison between artificial and natural aging protocols. Plant Physiol 1:620–641. https://doi.org/10.1104/pp.108.123141
Rajjou L, Duval M, Gallardo K, Catusse J, Bally J, Job C, Job D (2012) Seed germination and vigor. Annu Rev Plant Biol 63:507–533. https://doi.org/10.1146/annurev-arplant-042811-105550
Razem FA, Baron K, Hill RD (2006) Turning on gibberellin and abscisic acid signaling. Curr Opin Plant Biol 9:454–459. https://doi.org/10.1016/j.pbi.2006.07.007
Ribeiro RP, Costa LC, Medina EF, Araújo WL, Zsögön A, Ribeiro DM (2018) Ethylene coordinates seed germination behavior in response to low soil pH in Stylosanthes humilis. Plant Soil 425:87–100. https://doi.org/10.1007/s11104-018-3572-2
Saxena K, Kumar R, Sultana R (2010) Quality nutrition through pigeonpea-a review. Health 11:1335–1344. https://doi.org/10.4236/health.2010.211199
Shivashankar S, Sumathi M (2019) Rapid burst of ethylene evolution by premature seed: A warning sign for the onset of spongy tissue disorder in Alphonso mango fruit? J Biosci 44:133–152. https://doi.org/10.1007/s12038-019-9957-4
Silva LJD, Dias DCFDS, Sekita MC, Finger FL (2018) Lipid peroxidation and antioxidant enzymes of Jatropha curcas L. seeds stored at different maturity stages. Acta Scientiarum Agron 40:34978. https://doi.org/10.4025/actasciagron.v40i1.34978
Skibinski GA, Boyd L (2012) Ubiquitination is involved in secondary growth, not initial formation of polyglutamine protein aggregates in C. elegans. BMC Cell Biol 13:10–15. https://doi.org/10.1186/1471-2121-13-10
Song SQ, Fredlund KM, Moller IM (2001) Changes in low-molecular weight heat shock protein 22 of mitochondria during high-temperature accelerated ageing of Beta vulgaris L.seeds. Acta Phytophysiol Sin 27:73–80
Song SQ, Cheng HY, Long CL, Jiang XC (2005) A guide to seed biology research. Science Press, Beijing, pp 1–107
Sun TP (2008) Gibberellin metabolism, perception and signaling pathways in Arabidopsis. Arabidopsis Book 2008:e0103. https://doi.org/10.1199/tab.0103
Tatic M, Balesevic-Tubic S, Dordevic V, Nikolic Z, Dukic V, Vujakovic M, Cvijanovic G (2012) Soybean seed viability and changes of fatty acids content as affected by seed aging. Afr J Biotechnol 11:10310–10325
Timotijević GS, Milisavljević MD, Radović SR, Konstantinović MM, Maksimović VR (2010) Ubiquitous aspartic proteinase as an actor in the stress response in buckwheat. J Plant Physiol 167:61–68. https://doi.org/10.1016/j.jplph.2009.06.017
Ventura L, Donà M, Macovei A, Carbonera D, Buttafava A, Mondoni A, Rossi G, Balestrazzi A (2012) Understanding the molecular pathways associated with seed vigor. Plant Physiol Biochem 60:196–206. https://doi.org/10.1016/j.plaphy.2012.07.031
Verma P, Kaur H, Petla BP, Rao V, Saxena SC, Majee M (2013) Protein l-isoaspartyl methyltransferase2 is differentially expressed in chickpea and enhances seed vigor and longevity by reducing abnormal isoaspartyl accumulation predominantly in seed nuclear proteins. Plant Physiol 161:1141–1158. https://doi.org/10.1104/pp.112.206243
Wang T, Hou L, Jian H, Di F, Li J, Liu L (2018) Combined QTL mapping, physiological and transcriptomic analyses to identify candidate genes involved in Brassica napus seed aging. Mol Genet Genom 293:1421–1435. https://doi.org/10.1007/s00438-018-1468-8
Wei YD, Xu HB, Diao LR, Zhu YS, Xie HG, Cai QH, Wu FX, Wang ZH, Zhang JF, Xie HA (2015) Protein repair L-isoaspartyl methyltransferase 1 (PIMT1) in rice improves seed longevity by preserving embryo vigor and viability. Plant Mol Biol 89:475–492. https://doi.org/10.1007/s11103-015-0383-1
Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144:1240–1246. https://doi.org/10.1104/pp.107.100370
Wuddineh WA, Mazarei M, Zhang J, Poovaiah CR, Mann DGJ, Ziebell A, Sykes RW, Davis MF, Udvardi MK, Stewart CN Jr (2015) Identification and overexpression of gibberellin 2-oxidase (GA2ox) in switchgrass (Panicum virgatum L.) for improved plant architecture and reduced biomass recalcitrance. Plant Biotechnol J 13:636–647. https://doi.org/10.1111/pbi.12287
Yin X, He D, Gupta YPF (2015) Physiological and proteomic analyses on artificially aged Brassica napus seed. Front Plant Sci 6:121–132. https://doi.org/10.3389/fpls.2015.00112
Yu YL, Guo GF, Lv DW, Hu YK, Li JR, Li XH, Yan YM (2014) Transcriptome analysis during seed germination of elite Chinese bread wheat cultivar Jimai 20. BMC Plant Biol 14:20–38. https://doi.org/10.1186/1471-2229-14-20
Zabala G, Kour A, Vodkin LO (2020) Overexpression of an ethylene-forming ACC oxidase (ACO) gene precedes the minute hilum seed coat phenotype in glycine max. BMC Genom 21:716–735. https://doi.org/10.1186/s12864-020-07130-8
Zamyatnin AA (2015) Plant proteases involved in regulated cell death. Biochem Mosc 80:1701–1715. https://doi.org/10.1134/S0006297915130064
Zhang XY, Yang ZR, Hao LZ, Zhang FL, Zheng QL, Guo F, Zhai XJ (2017) Effect of salt stress on germination and oxygen processing of seed in Cynanchum thesiodes (Freyn). K Schum Acta Bot Boreal-Occident Sin 37:1166–1174. https://doi.org/10.7606/j.issn.1000-4025.2017.06.1166
Funding
This work was supported by a National Natural Science Foundations of China (311,760,081), Science Fund of Inner Mongolia of China (2019MS03055), Animal and Plant Breeding Special projects of Inner Mongolia Agricultural University (YZGC2017016), The Major Seed Industry Science and Technology Innovation Demonstration Project of the Autonomous Region Unveiled in Command, The Special Fund Project for the Transformation of Scientific and Technological Achievements of the Inner Mongolia Autonomous Region (2021CG0023), The Science and Technology Plan of the Inner Mongolia Autonomous Region (2021GG0084), The Major Science and Technology Project of the Inner Mongolia Autonomous Region (2021ZD0001), Major Science and Technology Project (2019ZD016).
Author information
Authors and Affiliations
Contributions
H, LZ, H, XM and Y, ZR supervised the experiment. Z, XY, G, F, Z, D, L, Z, N, KZ and W, PC performed the experiment work and prepared the figures and tables. Z, XY wrote the manuscript. Z, FL edited the final version of the manuscript.All authors have approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declared no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10681_2023_3259_MOESM1_ESM.docx
Supplementary file1 Figure S1 (A) Length distribution of Allium mongolicum unigenes; (B)All samples Pearson; (C)PCA plot analysis of all samples; (D) The number of DEGs after different storage times (1, 4, 8 years).
10681_2023_3259_MOESM3_ESM.docx
Supplementary file3 Figure S3 Histogram of gene ontology classification. The results have been summarized into three main GO categories: biological processes, cellular components and molecular function. The x-axis indicates the sub-categories, and the y-axis indicates the number of DEGs. (A) GO terms of S1 vs S4; (B) GO terms of S2 vs S4.
10681_2023_3259_MOESM4_ESM.docx
Supplementary file4 Figure S4 Heatmap of DEGs involved in pathways both in S1 vs S4 and S2 vs S4 (A) hormone; (B) signaling; (C) redox and secondary metabolism. The expression levels of DEGs in RPKM were compiled using Excel 2007, normalized using a log2 base method, and exported to the HemItoolkit 124. The heatmap was generated using default parameters. The expression levels are presented as different colours and values are defined in the scale bar.
10681_2023_3259_MOESM6_ESM.docx
Supplementary file6 Table S2 Basic information on transcriptome data from A. mongolicum seeds stored for 1, 4 and 8 year. (A) Summary of sequences analysis; (B) Statistics of assembly results; (C) Statistical tables for the four databases annotated.
10681_2023_3259_MOESM8_ESM.txt
Supplementary file8 Table S4 Differentially expressed genes involved in protein and energy metabolism in both S1 vs S4 and S2 vs S4
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, X., Guo, F., Huang, X. et al. Combined physiological and transcriptomic analyses to identify candidate genes involved in aging during storage of Allium mongolicum Regel. seeds. Euphytica 220, 14 (2024). https://doi.org/10.1007/s10681-023-03259-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03259-1