Abstract
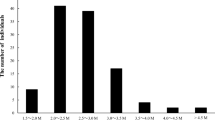
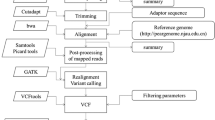
Almond (Prunus dulcis (Mill) D.A. Webb) is the only nut tree species in Rosaceae family and it is one of the most important nut crops all over the world. Cultivar breeding programs in almond take a long time due to its relatively long juvenile period. In the present study, we aimed (1) to construct high-density genetic linkage maps in two intra-specific almond populations, (2) to detect syntenic rates of the markers derived from ‘Gulcan-2 × Lauranne’ and ‘Guara × Nurlu’ F1 populations and (3) to identify synteny of the almond genome with peach, Japanese apricot and sweet cherry genomes. In this study, diversity arrays technology (DArT) and single nucleotide polymorphisms (SNP) markers were used for high-density genetic linkage map construction. In ‘Gulcan-2 × Lauranne’ population, the ‘Gulcan-2’ maternal linkage map included 3206 markers, and the total map length was 436.8 cM with 7.34 marker density (marker/cM), whereas the ‘Lauranne’ paternal genetic map contained 3308 markers, and the total map length was 486.4 cM with 6.80 marker density. In ‘Guara × Nurlu’ population, the ‘Guara’ maternal linkage map included 2430 markers, and total map length was 378.6 cM with 6.42 marker density (marker/cM), whereas the ‘Nurlu’ paternal genetic map contained 3,005 markers, and the total map length was 497.0 cM with 6.05 marker density. The highest synteny ratio was detected between almond and peach with 69.22%, while the lowest synteny ratio was detected between almond and sweet cherry with 54.75%. Synteny analysis revealed that high levels of the collinear gene pairs were detected with peach and sweet cherry genomes as pseudochromosomes in the same chromosome pairs. However, Japanese apricot had high synteny percentage with almond genome in different chromosomes. The genetic linkage maps based on DArT and SNP loci generated in this study will be very useful for future quantitative trait loci (QTL) analysis, synteny analysis of the almond genome with other Prunus species can be used for molecular breeding, and genetic studies in Prunus species.





Similar content being viewed by others
Data availability
All analyzed data during this study are included in this published article [and its supplementary information files]. LGs, loci, positions, significance, and segregations belonging to ‘Gulcan-2 × Lauranne’ and ‘Guara × Nurlu’ F1 almond populations are given in Supplementary File-1. The results of the blast of the loci in these populations to the almond genome have been shared in Supplementary File-2.
References
Alioto T, Alexiou KG, Bardil A et al (2020) Transposons played a major role in the diversification between the closely related almond and peach genomes: results from the almond genome sequence. Plant J. https://doi.org/10.1111/tpj.14538
Aranzana MJ, Decroocq V, Dirlewanger E, et al (2019) Prunus genetics and applications after de novo genome sequencing: achievements and prospects. Hortic. Res
Arús P, Messeguer R, Viruell M et al (1994) The European Prunus mapping project Progress in the almond linkage map. Euphytica. https://doi.org/10.1007/BF02551469
Ballester Sociasompany J, Socias I Company R, Arus P, Vicente MC (2001) Genetic mapping of a major gene delaying blooming time in almond. Plant Breed 56:5454. https://doi.org/10.1046/j.1439-0523.2001.00604.x
Calle A, Cai L, Iezzoni A, Wünsch A (2018) High-density linkage maps constructed in sweet cherry (Prunus avium L.) using cross- and self-pollination populations reveal chromosomal homozygosity in inbred families and non-syntenic regions with the peach genome. Tree Genet Genomes. https://doi.org/10.1007/s11295-018-1252-2
Çelik F, Balta MF, Ercişli S et al (2019) Tocopherol contents of almond genetic resources from eastern and western Turkey. Erwerbs-Obstbau. https://doi.org/10.1007/s10341-019-00425-5
Cruz VMV, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop Lesquerella and related species. PLoS ONE. https://doi.org/10.1371/journal.pone.0064062
Curtolo M, Cristofani-Yaly M, Gazaffi R et al (2017) QTL mapping for fruit quality in Citrus using DArTseq markers. BMC Genomics. https://doi.org/10.1186/s12864-017-3629-2
Curtolo M, Soratto TAT, Gazaffi R et al (2018) High-density linkage maps for Citrus sunki and Poncirus trifoliata using DArTseq markers. Tree Genet Genomes. https://doi.org/10.1007/s11295-017-1218-9
Delplancke M, Yazbek M, Arrigo N et al (2016) Combining conservative and variable markers to infer the evolutionary history of Prunus subgen. Amygdalus s.l. under domestication. Genet Resour Crop Evol 45:64. https://doi.org/10.1007/s10722-015-0242-6
Donoso JM, Picañol R, Serra O et al (2016) Exploring almond genetic variability useful for peach improvement: mapping major genes and QTLs in two interspecific almond × peach populations. Mol Breed. https://doi.org/10.1007/s11032-016-0441-7
Doyle J, Doyle J (1987) A rapid isolation procedure for small amounts of leaf tissue. Phytochem Bull. https://doi.org/10.2307/4119796
Elshire RJ, Glaubitz JC, Sun Q et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE. https://doi.org/10.1371/journal.pone.0019379
FAO (2022) FAOSTAT: Agriculture Data. Available online
Fernández Martí A, Font Forcada C, Kamali K et al (2015) Molecular analyses of evolution and population structure in a worldwide almond [Prunus dulcis (Mill.) D.A. Webb syn. P. amygdalus Batsch] pool assessed by microsatellite markers. Genet Resour Crop Evol. https://doi.org/10.1007/s10722-014-0146-x
Gardner KM, Brown P, Cooke TF et al (2014) Fast and cost-effective genetic mapping in apple using next-generation sequencing. G3 Genes Genomes, Genet. 4:1681–1687. https://doi.org/10.1534/g3.114.011023
Goonetilleke SN, March TJ, Wirthensohn MG et al (2018) Genotyping by sequencing in almond: SNP discovery, linkage mapping, and marker design. G3 Genes Genomes, Genet 8:161–172. https://doi.org/10.1534/g3.117.300376
Grasselly C, Crossa‐Raynaud P (1980) L’Amandier (Paris, France: GP Maisonneuve et Larosa)
Grattapaglia D, Sederoff R (1994) Genetic linkage maps of Eucalyptus grandis and Eucalyptus urophylla using a pseudo-testcross: mapping strategy and RAPD markers. Genetics. https://doi.org/10.1093/genetics/137.4.1121
Guajardo V, Hinrichsen P, Muñoz C (2015a) Breeding rootstocks for prunus species: Advances in genetic and genomics of peach and cherry as a model. Chil J Agric Res 75:17–27
Guajardo V, Solís S, Sagredo B et al (2015b) Construction of high density sweet cherry (Prunus avium L.) linkage maps using microsatellite markers and SNPs detected by genotyping-by-sequencing (GBS). PLoS ONE. https://doi.org/10.1371/journal.pone.0127750
Hasanbegovic J, Hadziabulic S, Kurtovic M et al (2021) Genetic characterization of almond (Prunus amygdalus L.) using microsatellite markers in the area of Adriatic Sea. Turkish J Agric for. https://doi.org/10.3906/tar-2103-82
Hong Y, Chen X, Liang X et al (2010) A SSR-based composite genetic linkage map for the cultivated peanut (Arachis hypogaea L.) genome. BMC Plant Biol
Jaccoud D, Peng K, Feinstein D, Kilian A (2001) Diversity arrays: a solid state technology for sequence information independent genotyping. Nucleic Acids Res. https://doi.org/10.1093/nar/29.4.e25
Joobeur T, Periam N, De Vicente MC et al (2000) Development of a second generation linkage map for almond using RAPD and SSR markers. Genome. https://doi.org/10.1139/g00-040
Kafkas S, Ozkan H, Sutyemez M (2005) DNA polymorphism and assessment of genetic relationships in walnut genotypes based on AFLP and SAMPL markers. J Am Soc Hortic Sci. https://doi.org/10.21273/jashs.130.4.585
Kilian A, Wenzl P, Huttner E, et al (2012) Diversity arrays technology: a generic genome profiling technology on open platforms BT - Data production and analysis in population genomics: Methods and Protocols. In: Methods in molecular biology
Klagges C, Campoy JA, Quero-García J et al (2013) Construction and comparative analyses of highly dense linkage maps of two sweet cherry intra-specific progenies of commercial cultivars. PLoS ONE. https://doi.org/10.1371/journal.pone.0054743
Kosambi DD (2016) The Estimation of Map Distances from Recombination Values. In: D.D. Kosambi
Liu D, Ma C, Hong W et al (2014) Construction and analysis of high-density linkage map using high-throughput sequencing data. PLoS ONE. https://doi.org/10.1371/journal.pone.0098855
Özdemir B, Okay FY, Sarikamiş G et al (2021) Crosstalk between flowering and cold tolerance genes in almonds (Amygdalus spp.). Turkish J Agric for. https://doi.org/10.3906/tar-2103-101
Paizila A, Karcı H, Ziya Motalebipour E et al (2022) Quantitative trait loci analysis for flower-related traits in almond (Prunus dulcis). Plant Breed. https://doi.org/10.1111/pbr.12986
Raman H, Raman R, McVittie B et al (2017) A major locus for manganese tolerance maps on chromosome A09 in a doubled haploid population of brassica napus L. Front Plant Sci. https://doi.org/10.3389/fpls.2017.01952
Salazar JA, Pacheco I, Shinya P et al (2017) Genotyping by sequencing for Snp-Based linkage analysis and identification of QTLs linked to fruit quality traits in Japanese plum (Prunus salicina lindl). Front Plant Sci. https://doi.org/10.3389/fpls.2017.00476
Sánchez-Pérez R, Ballester J, Dicenta F et al (2006) Comparison of SSR polymorphisms using automated capillary sequencers, and polyacrylamide and agarose gel electrophoresis: implications for the assessment of genetic diversity and relatedness in almond. Sci Hortic (amsterdam) 108:310–316
Sánchez-Pérez R, Pavan S, Mazzeo R et al (2019) Mutation of a bHLH transcription factor allowed almond domestication. Science 364(6445):1095–1098. https://doi.org/10.1126/science.aav8197
Shirasawa K, Isuzugawa K, Ikenaga M et al (2017) The genome sequence of sweet cherry (Prunus avium) for use in genomics-assisted breeding. DNA Res. https://doi.org/10.1093/dnares/dsx020
Tavassolian I, Rabiei G, Gregory D et al (2010) Construction of an almond linkage map in an Australian population Nonpareil × Lauranne. BMC Genomics. https://doi.org/10.1186/1471-2164-11-551
Tyrka M, Tyrka D, Wędzony M (2015) Genetic map of triticale integrating microsatellite DArT and SNP markers. PLoS ONE. https://doi.org/10.1371/journal.pone.0145714
Van Ooijen JW (2011) Multipoint maximum likelihood mapping in a full-sib family of an outbreeding species. Genet Res. https://doi.org/10.1017/S0016672311000279
Verde I, Jenkins J, Dondini L et al (2017) The Peach v20 release: High-resolution linkage mapping and deep resequencing improve chromosome-scale assembly and contiguity. BMC Genomics 454:65. https://doi.org/10.1186/s12864-017-3606-9
Viruel MA, Messeguer R, De Vicente MC et al (1995) A linkage map with RFLP and isozyme markers for almond. Theor Appl Genet 91:964–971
Voorrips RE (2002) Mapchart: Software for the graphical presentation of linkage maps and QTLs. J. Hered
Wang Y, Tang H, Debarry JD et al (2012) MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. https://doi.org/10.1093/nar/gkr1293
Wang J, Liu W, Zhu D et al (2020) Chromosome-scale genome assembly of sweet cherry (Prunus avium L.) cv. Tieton obtained using long-read and Hi-C sequencing. Hortic Res. https://doi.org/10.1038/s41438-020-00343-8
Ward JA, Bhangoo J, Fernández-Fernández F et al (2013) Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics. https://doi.org/10.1186/1471-2164-14-2
Wu SB, Tavassolian I, Rabiei G et al (2009) Mapping SNP-anchored genes using high-resolution melting analysis in almond. Mol Genet Genomics. https://doi.org/10.1007/s00438-009-0464-4
Yu Y, Fu J, Xu Y et al (2018) Genome re-sequencing reveals the evolutionary history of peach fruit edibility. Nat Commun. https://doi.org/10.1038/s41467-018-07744-3
Zhang Q, Chen W, Sun L et al (2012) The genome of Prunus mume. Nat Commun. https://doi.org/10.1038/ncomms2290
Funding
This research was financially supported by the Turkish General Directorate of Agricultural Research and Policies (TAGEM/14/ARGE/01) for financial support.
Author information
Authors and Affiliations
Contributions
HT and SK conceived and designed the experiments. HT, HK and JW performed most of the experiments and analyzed the data. HT, HK and SK wrote the manuscript. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interests.
Ethical approval and consent to participate
Not applicable.
Consent for publication
All authors have read and approved the manuscript for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tevfik, H., Karcı, H., Wai, J. et al. Construction of high density linkage maps in almond and synteny analysis with peach, Japanese apricot and sweet cherry genomes. Euphytica 219, 82 (2023). https://doi.org/10.1007/s10681-023-03210-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-023-03210-4