Abstract
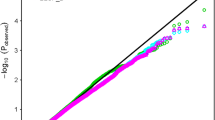
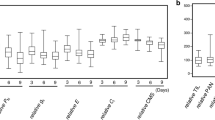
Seed germination is the first stage of the plant life cycle, and salinity is a major environmental stress for the inhibition of seed germination in plants. Hemp (Cannabis sativa. L) has high economic value and is widely used in textiles, building materials, food, health care, and so on. Unfortunately, research on the salt tolerance of hemp is very limited. In this study, 202 hemp accessions from 11 countries were used to conduct a genome-wide association study (GWAS) on salt tolerance at the germination stage. Based on 762,634 high-quality SNPs, 70 SNP loci were identified to be associated with salt tolerance-related traits (RGR-T1, RGR-T2, RSRL-T1, RSRL-T2, RSW-T1 and RSW-T2). Among the 70 SNPs, nine were further analyzed as candidate SNPs, and 55 genes were annotated within 100 kb upstream and downstream of the nine SNPs. Combined with RNA-seq data and qRT–PCR results, XM_030641043.1, XM_030641906.1, XM_030648362.1, XM_030648308.1 and XM_030646898.1 may play a key role in regulating salt tolerance of hemp during germination. To our knowledge, this is the first study of hemp GWAS, and the information obtained in this study will be useful for gene cloning and marker-assisted selection (MAS) of salt tolerance in hemp.





Similar content being viewed by others
References
Abbas MK, Ali AS, Hasan HH, Ghal RH (2013) Salt tolerance study of six cultivars of rice (Oryza sativa L.) during germination and early seedling growth. J Agric Sci 5:250–259
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Alonso JM, Ecker JR (2006) Moving forward in reverse: Genetic technologies to enable genome-wide phenomic screens in Arabidopsis. Nat Rev Genet 7:524–536
Basnet RK, Duwal A, Tiwari DN, Xiao D, Monakhos S, Bucher J, Visser RGF, Groot SPC, Bonnema G, Maliepaard C (2015) Quantitative trait locus analysis of seed germination and seedling vigor in Brassica rapa reveals QTL hotspots and epistatic interactions. Front Plant Sci 6:1032
Cheng X, Deng G, Su Y, Liu JJ, Yang Y, Du GH, Chen ZY, Liu FH (2016) Protein mechanisms in response to NaCl-stress of salt-tolerant and salt-sensitive industrial hemp based on iTRAQ technology. Ind Crops Prod 83:444–452
El-Hendawy S, Elshafei A, Al-Suhaibani N, Alotabi M, Hassan W, Dewir YH, Abdella K (2019) Assessment of the salt tolerance of wheat genotypes during the germination stage based on germination ability parameters and associated SSR markers. J Plant Interact 14:151–163
Flexas J, Bota J, Loreto F, Cornic G, Sharkey TD (2004) Diffusive and metabolic limitations to photosynthesis under drought and salinity in C3 plants. Plant Biol 6:269–279
Flint-Garcia SA, Thornsberry JM, Buckler IV (2003) Structure of linkage disequilibrium in plants. Annu Rev Plant Biol 54:357–374
Flowers TJ (2004) Improving crop salt tolerance. J Exp Bot 55:307–319
Foolad MR, Stoltz T, Dervinis C, Rodriguez RL, Jones RA (1997) Mapping QTLs conferring salt tolerance during germination in tomato by selective genotyping. Mol Breeding 3:269–277
Ginestet C (2011) ggplot2: elegant graphics for data analysis. J Roy Stat Soc Series A 174:245–246
Golldack D, Li C, Mohan H, Probst N (2014) Tolerance to drought and salt stress in plants: unraveling the signaling networks. Front Plant Sci 5:151–160
Gu Q, Ke H, Liu C, Lv X, Sun Z, Liu Z, Rong W, Yang J, Zhang Y, Wu L, Zhang G, Wang X, Ma Z (2021) A stable QTL qSalt-A04-1 contributes to salt tolerance in the cotton seed germination stage. Theor Appl Genet 134:2399–2410
Hanin M, Ebel C, Ngom M, Laplaze L, Masmoudi K (2016) New insights on plant salt tolerance mechanisms and their potential use for breeding. Front Plant Sci 7:1787
Hasegawa PM, Bressan RA, Zhu JK, Bohnert HJ (2000) Plant cellular and molecular responses to high salt. Ann Rev Plant Physiol Plant Mol Biol 51:463–499
Hazman M, Hause B, Eiche E, Nick P, Riemann M (2015) Increased tolerance to salt stress in OPDA-deficient rice ALLENE OXIDE CYCLASE mutants is linked to an increased ROS-scavenging activity. J Exp Bot 66:3339–3352
Hu H, Liu H, Liu F (2018) Seed germination of hemp (Cannabis sativa L.) cultivars responds differently to the stress of salt type and concentration. Ind Crops Prod 123:254–261
Hu H, Liu H, Du G, Yang F, Deng G, Yang Y, Liu F (2019) Fiber and seed type of hemp (Cannabis sativa L.) responded differently to salt-alkali stress in seedling growth and physiological indices. Ind Crops Prod 129:624–630
Huang X, Sang T, Zhao Q, Feng Q, Zhao Y, Li C, Zhu C, Lu T, Zhang Z, Li M, Fan D, Guo Y, Wang A, Wang L, Deng L, Li W, Lu Y, Weng Q, Liu K, Huang T, Zhou T, Jing Y, Li W, Lin Z, Buckler ES, Qian Q, Zhang QF, Li J, Han B (2010) Genome-wide association studies of 14 agronomic traits in rice landraces. Nat Genet 42:961
Huang X, Kurata N, Wei X, Wang ZX, Wang A, Zhao Q, Zhao Y, Liu K, Lu H, Li W, Guo Y, Lu Y, Zhou C, Fan D, Weng Q, Zhu C, Huang T, Zhang L, Wang Y, Feng L, Furuumi H, Kubo T, Miyabayashi T, Yuan X, Xu Q, Dong G, Zhan Q, Li C, Fujiyama A, Toyoda A, Lu T, Feng Q, Qian Q, Li J, Han B (2012) A map of rice genome variation reveals the origin of cultivated rice. Nature 490:497–501
Huang K, Shi Y, Pan G, Zhong Y, Sun Z, Niu J, Chen J, Luan M (2021) Genome-wide association analysis of fiber fineness and yield in ramie (Boehmeria nivea) using SLAF-seq. Euphytica 217:22
Jamil M, Lee KB, Jung KY, Lee DB, Han MS, Rha ES (2007) Salt stress inhibits germination and early seedling growth in cabbage (Brassica oleracea capitata L.). Pak J Biol Sci 10:910–914
Kan G, Zhang W, Yang W, Ma D, Zhang D, Hao D, Hu Z, Yu D (2015) Association mapping of soybean seed germination under salt stress. Mol Genet Genomics 290:2147–2162
Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E (2008) Efficient control of population structure in model organism association mapping. Genetics 178:1709–1723
Kang HM, Sul JH, Service SK, Zaitlen NA, Kong SY, Freimer NB, Sabatti C, Eskin E (2010) Variance component model to account for sample structure in genome-wide association studies. Nat Genet 42:348–354
Korte A, Farlow A (2013) The advantages and limitations of trait analysis with GWAS: a review. Plant Methods 9:29
Kronzucker HJ, Britto DT (2011) Sodium transport in plants: a critical review. New Phytol 189:54–81
Laverty KU, Stout JM, Sullivan MJ, Shah H, Gill N, Holbrook L, Deikus G, Sebra R, Hughes TR, Page JE, Bakel H (2018) A Physical and genetic map of Cannabis sativa identifies extensive rearrangement at the THC/CBD acid synthase locus. Genome Res 29:146–156
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079
Li Y, Zeng XF, Zhao YC, Li JR, Zhao DG (2017) Identification of a new rice low-tiller mutant and association analyses based on the SLAF-seq method. Plant Mol Biol Rep 35:72–82
Li D, Dossa K, Zhang Y, Wei X, Wang L, Zhang Y, Liu A, Zhou R, Zhang X (2018) GWAS uncovers differential genetic bases for drought and salt tolerances in sesame at the germination stage. Genes 9:87
Li W, Zhang H, Zeng Y, Xiang L, Lei Z, Huang Q, Li T, Shen F, Cheng Q (2020) A salt tolerance evaluation method for sunflower (Helianthus annuus L.) at the seed germination stage. Sci Rep 10:10626
Liu J, Qiao Q, Cheng X, Du G, Deng G, Zhao M, Liu F (2016a) Transcriptome differences between fiber-type and seed-type Cannabis sativa variety exposed to salinity. Physiol Mol Biol Plants 22:429–443
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016b) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12:e1005767
Liu L, Xia W, Li H, Zeng H, Wei B, Han S, Yin C, Villasuso AL (2018) Salinity inhibits rice seed germination by reducing α-amylase activity via decreased bioactive gibberellin content. Front Plant Sci 9:275–284
Luo Z, Szczepanek A, Abdel-Haleem H (2020) Genome-wide association study (GWAS) analysis of camelina seedling germination under salt stress condition. Agronomy 10:1444
Ma L, Zhou E, Huo N, Zhou R, Wang G, Jia J (2007) Genetic analysis of salt tolerance in a recombinant inbred population of wheat (Triticum aestivum L.). Euphytica 153:109–117
Mandozai A, Moussa AA, Zhang Q, Qu J, Du Y, Anwari G, Amin NA, Wang P (2021) Genome-wide association study of root and shoot related traits in spring soybean (Glycine max L.) at seedling stages using SLAF-Seq. Front Plant Sci 12:568995
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303
Munns R, James RA (2003) Screening methods for salinity tolerance: a case study with tetraploid wheat. Plant Soil 253:201–218
Munns R, Tester M (2008) Mechanisms of salinity tolerance. Ann Rev Plant Biol 59:651–681
Murray MG, Thompson WF (1980) Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res 8:4321–4326
Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D (2006) Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet 38:904–909
Ravelombola W, Shi A, Weng Y, Mou B, Motes D, Clark J, Chen P, Srivastava V, Qin J, Dong L, Yang W, Bhattarai G, Sugihara Y (2018) Association analysis of salt tolerance in cowpea (Vigna unguiculata (L.) Walp) at germination and seedling stages. Theor Appl Genet 131:79–91
Salentijn EMJ, Zhang QY, Amaducci S, Yang M, Trindade LM (2015) New developments in fiber hemp (Cannabis sativa L.) breeding. Ind Crops Prod 68:32–41
Shi Y, Gao L, Wu Z, Zhang X, Wang M, Zhang C, Zhang F, Zhou Y, Li Z (2017) Genome-wide association study of salt tolerance at the seed germination stage in rice. BMC Plant Biol 17:92
Song J, Wang B (2015) Using euhalophytes to understand salt tolerance and to develop saline agriculture: Suaeda salsa as a promising model. Ann Bot 115:541–553
Su J, Li L, Zhang C, Wang C, Gu L, Wang H, Wei H, Liu Q, Huang L, Yu S (2018) Genome-wide association study identified genetic variations and candidate genes for plant architecture component traits in Chinese upland cotton. Theor Appl Genet 131:1299–1314
Sun X, Liu D, Zhang X, Li W, Liu H, Hong W, Jiang C, Guan N, Ma C, Zeng H (2013) SLAF-seq: an efficient method of large-scale de novo SNP discovery and genotyping using high-throughput sequencing. PLoS ONE 8:e58700
Tsugane K, Kobayashi K, Niwa Y, Ohba Y, Wada K, Kobayashi H (1999) A recessive Arabidopsis mutant that grows photoautotrophically under salt stress shows enhanced active oxygen detoxification. Plant Cell 11:1195–1206
Turner SD (2014) qqman: An R package for visualizing GWAS results using QQ and manhattan plots. BioRxiv 005165
Vallejo AJ, Yanovsky MJ, Botto JF (2010) Germination variation in Arabidopsis thaliana accessions under moderate osmotic and salt stresses. Ann Bot 106:833–842
Wan H, Wei Y, Qian J, Gao Y, Wen J, Yi B, Ma C, Tu J, Fu T, Shen J (2018) Association mapping of salt tolerance traits at germination stage of rapeseed (Brassica napus L.). Euphytica 214:190
Wang Z, Wang J, Bao Y, Wu Y, Zhang H (2011) Quantitative trait loci controlling rice seed germination under salt stress. Euphytica 178:297–307
Wang Q, Tang JL, Han B, Huang XH (2020) Advances in genome-wide association studies of complex traits in rice. Theor Appl Genet 133:1415–1425
Wei X, Liu K, Zhang Y, Feng Q, Wang L, Zhao Y, Li D, Zhao Q, Zhu X, Zhu X, Li W, Fan D, Gao Y, Lu Y, Zhang X, Tang X, Zhou C, Zhu C, Liu L, Zhong R, Tian Q, Wen Z, Weng Q, Han B, Huang X, Zhang X (2015) Genetic discovery for oil production and quality in sesame. Nat Commun 26:8609
Wu H, Guo J, Wang C, Li K, Zhang X, Yang Z, Li M, Wang B (2019) An Effective Screening Method and a Reliable Screening Trait for Salt Tolerance of Brassica napus at the Germination Stage. Front Plant Sci 10:530
Xiao X, Sha L, Fan X, Zhou Y (2012) Salt tolerance on seed germination of five leymus species. World J Agr Sci 8:585–589
Xie D, Dai Z, Yang Z, Tang Q, Sun J, Yang X, Song X, Lu Y, Zhao D, Zhang L, Su J (2018) Genomic variations and association study of agronomic traits in flax. BMC Genomics 19:512
Yang Y, Guo Y (2018) Elucidating the molecular mechanisms mediating plant salt-stress responses. New Phytol 217:523–539
Yang J, Yang M, Su L, Zhou D, Huang C, Wang H, Guo T, Chen Z (2020) Genome-wide association study reveals novel genetic loci contributing to cold tolerance at the germination stage in indica rice. Plant Sci 301:110669
Yep B, Gale NV, Zheng Y (2020) Aquaponic and hydroponic solutions modulate NaCl-induced stress in drug-type Cannabis sativa L. Front Plant Sci 11:1169
Yu J, Zao W, He Q, Kim TS, Park YJ (2017) Genome-wide association study and gene set analysis for understanding candidate genes involved in salt tolerance at the rice seedling stage. Mol Genet Genomics 292:1391–1403
Yu J, Zhao W, Tong W, He Q, Yoon MY, Li FP, Choi B, Heo EB, Kim KW, Park YJ (2018) A genome-wide association study reveals candidate genes related to salt tolerance in rice (Oryza sativa) at the germination stage. Int J Mol Sci 19:3145
Yu LX, Liu X, Boge W, Liu XP (2016) Genome-wide association study identifies loci for salt tolerance during germination in autotetraploid alfalfa (Medicago sativa L.) using Genotyping-by-Sequencing. Front Plant Sci 7:956
Yuan Y, Xing H, Zeng W, Xu J, Mao L, Wang L, Feng W, Tao J, Wang H, Zhang H, Wang Q, Zhang G, Song X, Sun XZ (2019) Genome-wide association and differential expression analysis of salt tolerance in Gossypium hirsutum L at the germination stage. BMC Plant Biol 19:394
Zhang H, Irving LJ, Matthew MC, Zhou D, Kemp P, (2010a) The effects of salinity and osmotic stress on barley germination rate: sodium as an osmotic regulator. Ann Bot 106:1027–1035
Zhang Z, Ersoz E, Lai CQ, Todhunter RJ, Tiwari HK, Gore MA, Bradbury PJ, Yu J, Arnett DK, Ordovas JM, Buckler ES (2010b) Mixed linear model approach adapted for genome-wide association studies. Nat Genet 42:355
Zhou X, Stephens M (2012) Genome-wide efficient mixed-model analysis for association studies. Nat Genet 44:821–826
Acknowledgements
This work was supported by the Special Construction of Modern Agricultural Industrial Technology System (CARS-16-E01), Protection and Utilization of Crop Germplasm Resources (2016NWB044), National Science and Technology Resource Sharing Service Platform Project (NCGRC-2020-15).
Funding
The authors have not disclosed any funding.
Author information
Authors and Affiliations
Contributions
JGS, JS and ZGD designed the experiments. JQC and GCX contributed to the phenotypic evaluation. YY and XYZ contributed to the genotyping. JS wrote the manuscript. All authors reviewed and approved this submission.
Corresponding authors
Ethics declarations
Conflict of interest
Authors have no competing interests to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Sun, J., Chen, J., Zhang, X. et al. Genome-wide association study of salt tolerance at the germination stage in hemp. Euphytica 219, 5 (2023). https://doi.org/10.1007/s10681-022-03129-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-03129-2