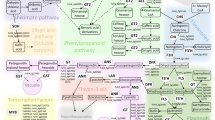
Abstract
Resveratrol, a polyphenolic compound, is related with stress resistance or tolerance in plants and highly beneficial to human health. Peanut (Arachis hypogaea L.) is not only an important oilseed crop worldwide but also a key source of resveratrol from dietary food. In this study, a peanut recombinant inbred line (RIL) population consisting of 166 lines derived from a cross combination with a high resveratrol variety ICGV86699 and a normal resveratrol variety Zhonghua 5 as parents was assessed for resveratrol content across three environments. Broad phenotypic variation of resveratrol content ranging from 76.75 to 1617.00 µg/kg was observed among the RILs. From phenotyping resveratrol content and other traits, three elite lines (QT079, QT135, and QT141) were identified with resveratrol level as high as ICGV86699 while their seeds weights were around 1.5 times of ICGV8669. Nine additive QTLs with phenotypic variation explained (PVE) ranging from 6.66 to 11.33% were detected for resveratrol contnet, among which, qRESA05.3 with 7.88–10.91% PVE was repeatedly detected. Furthermore, an InDel marker (InDel-A05-8,713,397) based on qRESA05.3 could be detected in both the RILs and selected diverse peanut germplasm accessions possessing “aa” genotype, and the average resveratrol content of these “aa” accessions was significantly higher than that in the genotypes possessing “AA”. The favorable allele was found to raise the resveratrol content by 292.15 ± 32.05 µg/kg in the diverse germplasm panel. This is the first report of stable marker for peanut resveratrol, which would be helpful for further fine-mapping the locus and developing marker-assisted selection strategy for high resveratrol peanut.





Similar content being viewed by others
References
Baring MR, Wilson JN, Burow MD, Simpson CE, Ayers JL, Cason JM (2013) Variability of total oil content in peanut across the state of texas. J Crop Improv 27:125–136
Bertioli DJ, Jenkins J, Clevenger J, Dudchenko O, Gao DY, Seijo G, Leal-Bertioli SCM, Ren LH, Farmer AD, Pandey MK, Samoluk SS, Abernathy B, Agarwal G, Ballen-Taborda C, Cameron C, Campbell J, Chavarro C, Chitikineni A, Chu Y, Dash S et al (2019) The genome sequence of segmental allotetraploid peanut Arachis hypogaea. Nat Genet 51:877–884
Chen WG, Jiao YQ, Cheng LQ, Huang L, Liao BS, Tang M, Ren XP, Zhou XJ, Chen YN, Jiang HF (2016) Quantitative trait locus analysis for pod- and kernel-related traits in the cultivated peanut (Arachis hypogaea L.). BMC Genet 17:25
Chung IM, Park MR, Chun JC, Yun SJ (2003) Resveratrol accumulation and resveratrol synthase gene expression in response to abiotic stresses and hormones in peanut plants. Plant Sci 164:103–109
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
FAOSTAT (2020) Food and Agriculture Organization of the United Nations. http://faostat3.fao.org
Fouad MA, Agha AM, Merzabani MA, Shouman SA (2013) Resveratrol inhibits proliferation, angiogenesis and induces apoptosis in colon cancer cells. Hum Exp Toxicol 32:1067–1080
Gupta PK, Varshney RK (2013) QTL mapping: methodology and applications in cereal breeding. Springer, Netherlands
Jang M, Cai L, Udeani GO, Slowing KV, Thomas CF, Beecher CWW, Fong HHS, Farnsworth NR, Kinghorn AD, Mehta RG, Moon RC, Pezzuto JM (1997) Cancer chemopreventive activity of resveratrol, a natural product derived from grapes. Science 275:218–220
Jeandet P, Douillt-Breuil AC, Bessis R, Debord S, Sbaghi M, Adrian M (2002) Phytoalexins from the vitaceae: biosynthesis, phytoalexin gene expression in transgenic plants, antifungal activity, and metabolism. J Agr Food Chem 50:2731–2741
Kpl B, Kosmeder JW, Pezzuto JM, Fremont L (2002) Biological effects of resveratrol. Antioxid Redox Sign 3:1041–1064
Lee SS, Lee SM, Kim M, Chun J, Cheong YK, Lee J (2004) Analysis of trans-resveratrol in peanuts and peanut butters consumed in Korea. Food Res Int 37:247–251
Leonard SS, Xia C, Jiang BH, Stinefelt B, Klandorf H, Harris GK, Shi XL (2003) Resveratrol scavenges reactive oxygen species and effects radical-induced cellular responses. Biochem Biophys Res Commun 309:1017–1026
Liu N, Guo JB, Zhou XJ, Wu B, Huang L, Luo HY, Chen YN, Chen WG, Lei Y, Huang Y, Liao BS, Jiang HF (2020) High-resolution mapping of a major and consensus quantitative trait locus for oil content to a ~ 0.8-Mb region on chromosome A08 in peanut (Arachis hypogaea L.). Theor Appl Genet 133:37–49
Luo HY, Guo JB, Ren XP, Chen WG, Huang L, Zhou XJ, Chen YN, Liu N, Xiong F, Lei Y, Liao BS, Jiang HF (2018) Chromosomes A07 and A05 associated with stable and major QTLs for pod weight and size in cultivated peanut (Arachis hypogaea L.). Theor Appl Genet 131:267–282
Luo HY, Pandey MK, Zhi Y, Zhang H, Xu SL, Guo JB, Wu B, Chen HW, Ren XP, Zhou XJ, Chen YN, Chen WG, Huang L, Liu N, Sudini HK, Varshney RK, Lei Y, Liao BS, Jiang HF (2020) Discovery of two novel and adjacent QTLs on chromosome B02 controlling resistance against bacterial wilt in peanut variety Zhonghua 6. Theor Appl Genet 133:1133–1148
Luo HY, Guo JB, Yu BL, Chen WG, Zhang H, Zhou XJ, Chen YN, Huang L, Liu N, Ren XP, Yan LY, Huai DX, Lei Y, Liao BS, Jiang HF (2021) Construction of ddRADseq-based High-density genetic map and identification of quantitative trait loci for trans-resveratrol content in peanut seeds. Front Plant Sci 12:644402
Meng L, Li H, Zhang L, Wang J (2015) QTL IciMapping: integrated software for genetic linkage map construction and quantitative trait locus mapping in biparental populations. Crop J 3:269–283
Meredith Z, Alfred A (2003) Peanuts: a source of medically important resveratrol. Nat Prod Radiance 2:182–189
Mondal S, Badigannavar AM (2019) Identification of major consensus QTLs for seed size and minor QTLs for pod traits in cultivated groundnut (Arachis hypogaea L.). 3 Biotech 9:1–9
Ogaki Y, Sagawa I (2003) Trans-Resveratrol content in seeds and seed coats of Japanese peanut cultivars and that in peanut products. Nippon Shokuhin Kogyo Gakkaishi 50:570–573
Okamoto H, Matsukawa T, Doi S, Tsunoda T, Sawata Y, Naemura M, Ohnuki K, Shirasawa S, Kotake Y (2018) A novel resveratrol derivative selectively inhibits the proliferation of colorectal cancer cells with KRAS mutation. Mol Cell Biochem 442:39–45
Reddy LJ, Nigam SN, Singh AK, Moss JP, Subrahmanyam P, McDonald D, Reddy AGS (1996) Registration of ICGV 86699 peanut germplasm line with multiple disease and insect resistance. Crop Sci 36:821
Sales JM, Resurreccion AVA (2014) Resveratrol in peanuts. Crit Rev Food Sci 54:734–770
Sanders TH, Mcmichael RW, Hendrix KW (2000) Occurrence of resveratrol in edible peanuts. J Agr Food Chem 48:1243–1246
Sarvamangala C, Gowda M, Varshney RK (2011) Identification of quantitative trait loci for protein content, oil content and oil quality for groundnut (Arachis hypogaea L.). Field Crop Res 122:49–59
Shomura Y, Torayama I, Suh DY, Xiang T, Kita A, Sankawa U, Miki K (2005) Crystal structure of stilbene synthase from Arachis hypogaea. Proteins 60:803–806
Varshney RK, Murali MS, Gaur PM, Gangarao NVPR, Pandey MK, Bohra A, Sawargaonkar SL, Chitikineni A, Kimurto PK, Janila P, Saxena KB, Fikre A, Sharma M, Rathore A, Pratap A, Tripathi S, Datta S, Chaturvedi SK, Mallikarjuna N, Anuradha G (2013) Achievements and prospects of genomics-assisted breeding in three legume crops of the semi-arid tropics. Biotechnol Adv 31:1120–1134
Wang S, Basten C (2012) Windows QTL Cartographer 2.5. Department of Statistics, North Carolina State University, Raleigh, NC
Wang ML, Pittman RN (2009) Resveratrol content in seeds of peanut germplasm quantified by HPLC. Plant Genet Resour 7:80–83
Wang ML, Chen CY, Tonnis B, Barkley NA, Pinnow DL, Pittman RN, Davis J, Holbrook CC, Stalker HT, Pederson GA (2013) Oil, fatty acid, flavonoid, and resveratrol content variability and FAD2A functional SNP genotypes in the U.S. peanut mini-core collection. J Agr Food Chem 61:2875–2882
Xu SL, Luo HY, Chen HW, Guo JB, Yu BL, Zhang H, Li WT, Chen WG, Zhou XJ, Huang L, Liu N, Lei Y, Liao BS, Jiang HF (2020) Optimization of extraction of total trans-resveratrol from peanut seeds and its determination by HPLC. J Sep Sci 43:1024–1031
Yu BL, Jiang HF, Pandey MK, Huang L, Huai DX, Zhou XJ, Kang YP, Varshney RK, Sudini HK, Ren XP, Luo HY, Liu N, Chen WG, Guo JB, Li WT, Ding YB, Jiang YF, Lei Y, Liao BS (2020) Identification of two novel peanut genotypes resistant to aflatoxin production and their SNP markers associated with resistance. Toxins 12:156
Zhou XJ, Xia YL, Ren XP, Chen YN, Huang L, Huang SM, Liao BS, Lei Y, Yan LY, Jiang HF (2014) Construction of a SNP-based genetic linkage map in cultivated peanut based on large scale marker development using next-generation double-digest restriction-site-associated DNA sequencing (ddRADseq). BMC Genom 15:351
Funding
This work was supported by the National Natural Science Foundation of China (31601340, 31801403, and 31871666), the Natural Science Foundation of Hubei Province (2017CFB540), the Central Public-interest Scientific Institution Basal Research Fund (1610172019008), the National Program for Crop Germplasm Protection of China (2019NWB033), and the Agricultural Science and Technology Innovation Program of Chinese Academy of Agricultural Sciences.
Author information
Authors and Affiliations
Contributions
JG, NL and HJ conceived and designed the research. XZ and WC developed the RIL population. LH, WL and HC planted the materials and conducted field management. JG, HC and BW performed the measurement of resveratrol content. JG analyzed the data and wrote the manuscript. NL, HL, DH, YL, BL and HJ revised the manuscript and improved the English writing. All the authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
The experiments were performed in compliance with the current laws of China.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Guo, J., Chen, H., Liu, N. et al. Identification and validation of a major locus with linked marker for resveratrol content in culitivated peanut. Euphytica 218, 15 (2022). https://doi.org/10.1007/s10681-022-02969-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-022-02969-2