Abstract
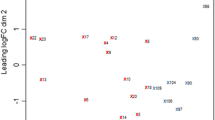
Sugarcane is a highly productive, first generation biofuel feedstock, known for its remarkable efficiency in accumulating biomass. Hormones are important regulators for many biological processes in plants, especially in plant development and plant growth, which are crucial for plant biomass traits. To understand how hormones regulatory mechanisms contribute to sugarcane lignocellulose yield, we studied the transgressive segregation on biomass yield in the F2 population derived from a cross between Saccharum officinarum ‘LA Purple’ and Saccharum robustum ‘MOL5829’. Gene expression profiling was used to detect genes involved in three important hormone-related pathways, auxin, ethylene and gibberellin, to find out how they are differently regulated between the extreme segregants of high and low biomass yield groups. We identified seventeen differentially expressed genes in auxin, one in ethylene and one in gibberellin related signaling and biosynthesis pathways, which could potentially regulate biomass yield. Differentially expressed genes, PIF3 and EIL5, involved in gibberellin and ethylene pathway could play an important role in biomass accumulation. These plant hormone-related genes could serve as candidate genes in genetic modification and breeding programs to develop high yielding energy cane.




Similar content being viewed by others
References
Abel S, Oeller PW, Theologis A (1994) Early auxin-induced genes encode short-lived nuclear proteins. Proc Natl Acad Sci USA 91(1):326–330
Abeles FB, Morgan PW, Saltveit ME (1992) Ethylene in plant biology. Academic Press, San Diego
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc
Arruda P (2011) Perspective of the sugarcane industry in Brazil. Trop Plant Biol 4(1):3–8. https://doi.org/10.1007/s12042-011-9074-5
Audran-Delalande C, Bassa C, Mila I, Regad F, Zouine M, Bouzayen M (2012) Genome-wide identification, functional analysis and expression profiling of the Aux/IAA gene family in tomato. Plant Cell Physiol 53(4):659–672. https://doi.org/10.1093/pcp/pcs022
Bandurski RS, Cohen JD, Slovin JP, Reinecke DM (1995) Auxin Biosynthesis and Metabolism
Biemelt S, Tschiersch H, Sonnewald U (2004) Impact of altered gibberellin metabolism on biomass accumulation, lignin biosynthesis, and photosynthesis in transgenic tobacco plants. Plant Physiol 135(1):254–265. https://doi.org/10.1104/pp.103.036988
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30(15):2114–2120. https://doi.org/10.1093/bioinformatics/btu170
Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71(4):684–697. https://doi.org/10.1111/j.1365-313X.2012.05024.x
Chao Q, Rothenberg M, Solano R, Roman G, Terzaghi W, Ecker JR (1997) Activation of the ethylene gas response pathway in Arabidopsis by the nuclear protein ETHYLENE-INSENSITIVE3 and related proteins. Cell 89(7):1133–1144
Chen JC, Chou C-C (1993) Cane sugar handbook: a manual for cane sugar manufacturers and their chemists, 12th edn. Wiley, New York
Chen G, Alexander L, Grierson D (2004) Constitutive expression of EIL-like transcription factor partially restores ripening in the ethylene-insensitive Nr tomato mutant. J Exp Bot 55(402):1491–1497. https://doi.org/10.1093/jxb/erh168
Chen Y, Hao X, Cao J (2014) Small auxin upregulated RNA (SAUR) gene family in maize: identification, evolution, and its phylogenetic comparison with Arabidopsis, rice, and sorghum. J Integr Plant Biol 56(2):133–150. https://doi.org/10.1111/jipb.12127
Choe S, Fujioka S, Noguchi T, Takatsuto S, Yoshida S, Feldmann KA (2001) Overexpression of DWARF4 in the brassinosteroid biosynthetic pathway results in increased vegetative growth and seed yield in Arabidopsis. Plant J 26(6):573–582
Cunha CP, Roberto GG, Vicentini R, Lembke CG, Souza GM, Ribeiro RV, Machado EC, Lagoa AMMA, Menossi M (2017) Ethylene-induced transcriptional and hormonal responses at the onset of sugarcane ripening. Sci Rep. https://doi.org/10.1038/Srep43364
Daviere JM, Achard P (2013) Gibberellin signaling in plants. Development 140(6):1147–1151. https://doi.org/10.1242/dev.087650
Dayan J, Schwarzkopf M, Avni A, Aloni R (2010) Enhancing plant growth and fiber production by silencing GA 2-oxidase. Plant Biotechnol J 8(4):425–435. https://doi.org/10.1111/j.1467-7652.2009.00480.x
Do PT, De Tar JR, Lee H, Folta MK, Zhang ZJ (2016) Expression of ZmGA20ox cDNA alters plant morphology and increases biomass production of switchgrass (Panicum virgatum L.). Plant Biotechnol J 14(7):1532–1540. https://doi.org/10.1111/pbi.12514
Dong X, Hong Z, Sivaramakrishnan M, Mahfouz M, Verma DP (2005) Callose synthase (CalS5) is required for exine formation during microgametogenesis and for pollen viability in Arabidopsis. Plant J 42(3):315–328. https://doi.org/10.1111/j.1365-313X.2005.02379.x
Ellis CM, Nagpal P, Young JC, Hagen G, Guilfoyle TJ, Reed JW (2005) AUXIN RESPONSE FACTOR1 and AUXIN RESPONSE FACTOR2 regulate senescence and floral organ abscission in Arabidopsis thaliana. Development 132(20):4563–4574. https://doi.org/10.1242/dev.02012
Energy OoEEaR, Bioenergy Technologies Office (2016) 2016 BILLION-TON REPORT. http://energy.gov/sites/prod/files/2016/08/f33/BillionTon_Report_2016_8.18.2016.pdf. Accessed 27 Nov 2016
Eriksson ME, Israelsson M, Olsson O, Moritz T (2000) Increased gibberellin biosynthesis in transgenic trees promotes growth, biomass production and xylem fiber length. Nat Biotechnol 18(7):784–788. https://doi.org/10.1038/77355
Fendrych M, Leung J, Friml J (2016) TIR1/AFB-Aux/IAA auxin perception mediates rapid cell wall acidification and growth of Arabidopsis hypocotyls. eLife. https://doi.org/10.7554/eLife.19048
Franco AR, Gee MA, Guilfoyle TJ (1990) Induction and superinduction of auxin-responsive mRNAs with auxin and protein synthesis inhibitors. J Biol Chem 265(26):15845–15849
Gray WM (2004) Hormonal regulation of plant growth and development. PLoS Biol 2(9):1270–1273. https://doi.org/10.1371/journal.pbio.0020311
Guo H, Ecker JR (2004) The ethylene signaling pathway: new insights. Curr Opin Plant Biol 7(1):40–49
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49(3–4):373–385
Jeon HW, Cho JS, Park EJ, Han KH, Choi YI, Ko JH (2016) Developing xylem-preferential expression of PdGA20ox1, a gibberellin 20-oxidase 1 from Pinus densiflora, improves woody biomass production in a hybrid poplar. Plant Biotechnol J 14(4):1161–1170. https://doi.org/10.1111/pbi.12484
Kant S, Bi YM, Zhu T, Rothstein SJ (2009) SAUR39, a small auxin-up RNA gene, acts as a negative regulator of auxin synthesis and transport in rice. Plant Physiol 151(2):691–701. https://doi.org/10.1104/pp.109.143875
Kosugi S, Ohashi Y (2000) Cloning and DNA-binding properties of a tobacco Ethylene-Insensitive3 (EIN3) homolog. Nucleic Acids Res 28(4):960–967
Lam E, Shine J, Da Silva J, Lawton M, Bonos S, Calvino M, Carrer H, Silva-Filho MC, Glynn N, Helsel Z, Ma J, Richard E, Souza GM, Ming R (2009) Improving sugarcane for biofuel: engineering for an even better feedstock. GCB Bioenergy 1(3):251–255. https://doi.org/10.1111/j.1757-1707.2009.01016.x
Lee JH, Kim WT (2003) Molecular and biochemical characterization of VR-EILs encoding mung bean ETHYLENE INSENSITIVE3-LIKE proteins. Plant Physiol 132(3):1475–1488
Leivar P, Monte E (2014) PIFs: systems integrators in plant development. Plant Cell 26(1):56–78. https://doi.org/10.1105/tpc.113.120857
Li ZG, Chen HW, Li QT, Tao JJ, Bian XH, Ma B, Zhang WK, Chen SY, Zhang JS (2015) Three SAUR proteins SAUR76, SAUR77 and SAUR78 promote plant growth in Arabidopsis. Sci Rep. https://doi.org/10.1038/Srep12477
Liscum E, Reed JW (2002) Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol 49(3–4):387–400
Ludwig-Muller J, Julke S, Bierfreund NM, Decker EL, Reski R (2009) Moss (Physcomitrella patens) GH3 proteins act in auxin homeostasis. New Phytol 181(2):323–338. https://doi.org/10.1111/j.1469-8137.2008.02677.x
Nakazawa M, Yabe N, Ichikawa T, Yamamoto YY, Yoshizumi T, Hasunuma K, Matsui M (2001) DFL1, an auxin-responsive GH3 gene homologue, negatively regulates shoot cell elongation and lateral root formation, and positively regulates the light response of hypocotyl length. Plant J 25(2):213–221
Nishikawa S, Zinkl GM, Swanson RJ, Maruyama D, Preuss D (2005) Callose (beta-1,3 glucan) is essential for Arabidopsis pollen wall patterning, but not tube growth. BMC Plant Biol 5:22. https://doi.org/10.1186/1471-2229-5-22
Pacala S, Socolow R (2004) Stabilization wedges: solving the climate problem for the next 50 years with current technologies. Science 305(5686):968–972. https://doi.org/10.1126/science.1100103
Paponov IA, Paponov M, Teale W, Menges M, Chakrabortee S, Murray JA, Palme K (2008) Comprehensive transcriptome analysis of auxin responses in Arabidopsis. Mol Plant 1(2):321–337. https://doi.org/10.1093/mp/ssm021
Pennington NL, Baker CW (1990) Sugar: user’s guide to sucrose. Springer Science & Business Media, New York
Qin X, Liu JH, Zhao WS, Chen XJ, Guo ZJ, Peng YL (2013) Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol Plant Microbe Interact 26(2):227–239. https://doi.org/10.1094/MPMI-05-12-0138-R
Ragauskas AJ, Williams CK, Davison BH, Britovsek G, Cairney J, Eckert CA, Frederick WJ Jr, Hallett JP, Leak DJ, Liotta CL, Mielenz JR, Murphy R, Templer R, Tschaplinski T (2006) The path forward for biofuels and biomaterials. Science 311(5760):484–489. https://doi.org/10.1126/science.1114736
Reinhardt D, Pesce ER, Stieger P, Mandel T, Baltensperger K, Bennett M, Traas J, Friml J, Kuhlemeier C (2003) Regulation of phyllotaxis by polar auxin transport. Nature 426(6964):255–260. https://doi.org/10.1038/nature02081
Rieu I, Mariani C, Weterings K (2003) Expression analysis of five tobacco EIN3 family members in relation to tissue-specific ethylene responses. J Exp Bot 54(391):2239–2244. https://doi.org/10.1093/jxb/erg240
Sahni S, Prasad BD, Liu Q, Grbic V, Sharpe A, Singh SP, Krishna P (2016) Overexpression of the brassinosteroid biosynthetic gene DWF4 in Brassica napus simultaneously increases seed yield and stress tolerance. Sci Rep 6:28298. https://doi.org/10.1038/srep28298
Searchinger T, Heimlich R, Houghton RA, Dong FX, Elobeid A, Fabiosa J, Tokgoz S, Hayes D, Yu TH (2008) Use of US croplands for biofuels increases greenhouse gases through emissions from land-use change. Science 319(5867):1238–1240. https://doi.org/10.1126/science.1151861
Shi ZH, Zhang C, Xu XF, Zhu J, Zhou Q, Ma LJ, Niu J, Yang ZN (2015) Overexpression of AtTTP affects ARF17 expression and leads to male sterility in Arabidopsis. PLoS ONE 10(3):e0117317. https://doi.org/10.1371/journal.pone.0117317
Shuai B, Reynaga-Pena CG, Springer PS (2002) The lateral organ boundaries gene defines a novel, plant-specific gene family. Plant Physiol 129(2):747–761. https://doi.org/10.1104/pp.010926
Singh VK, Jain M, Garg R (2014) Genome-wide analysis and expression profiling suggest diverse roles of GH3 genes during development and abiotic stress responses in legumes. Front Plant Sci 5:789. https://doi.org/10.3389/fpls.2014.00789
Solano R, Stepanova A, Chao Q, Ecker JR (1998) Nuclear events in ethylene signaling: a transcriptional cascade mediated by ETHYLENE-INSENSITIVE3 and ETHYLENE-RESPONSE-FACTOR1. Genes Dev 12(23):3703–3714
Somerville C, Youngs H, Taylor C, Davis SC, Long SP (2010) Feedstocks for lignocellulosic biofuels. Science 329(5993):790–792. https://doi.org/10.1126/science.1189268
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inze D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70(6):978–990. https://doi.org/10.1111/j.1365-313X.2012.04946.x
Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26(5):2129–2142. https://doi.org/10.1105/tpc.114.126037
Sticklen MB (2008) Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet 9(6):433–443. https://doi.org/10.1038/nrg2336
Tabashnik BE (2010) Plant science. Communal benefits of transgenic corn. Science 330(6001):189–190. https://doi.org/10.1126/science.1196864
Takase T, Nakazawa M, Ishikawa A, Kawashima M, Ichikawa T, Takahashi N, Shimada H, Manabe K, Matsui M (2004) ydk1-D, an auxin-responsive GH3 mutant that is involved in hypocotyl and root elongation. Plant J 37(4):471–483. https://doi.org/10.1046/j.1365-313X.2003.01973.x
Tatematsu K, Kumagai S, Muto H, Sato A, Watahiki MK, Harper RM, Liscum E, Yamamoto KT (2004) MASSUGU2 encodes Aux/IAA19, an auxin-regulated protein that functions together with the transcriptional activator NPH4/ARF7 to regulate differential growth responses of hypocotyl and formation of lateral roots in Arabidopsis thaliana. Plant Cell 16(2):379–393. https://doi.org/10.1105/tpc.108630
Tieman DM, Ciardi JA, Taylor MG, Klee HJ (2001) Members of the tomato LeEIL (EIN3-like) gene family are functionally redundant and regulate ethylene responses throughout plant development. Plant J 26(1):47–58
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13(12):2809–2822
Ulmasov T, Hagen G, Guilfoyle TJ (1999) Activation and repression of transcription by auxin-response factors. Proc Natl Acad Sci USA 96(10):5844–5849. https://doi.org/10.1073/Pnas.96.10.5844
Voorend W, Nelissen H, Vanholme R, De Vliegher A, Van Breusegem F, Boerjan W, Roldan-Ruiz I, Muylle H, Inze D (2016) Overexpression of GA20-OXIDASE1 impacts plant height, biomass allocation and saccharification efficiency in maize. Plant Biotechnol J 14(3):997–1007. https://doi.org/10.1111/pbi.12458
Wai CM, Zhang J, Jones TC, Nagai C, Ming R (2017) Cell wall metabolism and hexose allocation contribute to biomass accumulation in high yielding extreme segregants of a Saccharum interspecific F2 population. BMC Genomics 18(1):773. https://doi.org/10.1186/s12864-017-4158-8
Wang S, Bai Y, Shen C, Wu Y, Zhang S, Jiang D, Guilfoyle TJ, Chen M, Qi Y (2010) Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct Integr Genomics 10(4):533–546. https://doi.org/10.1007/s10142-010-0174-3
Willige BC, Ghosh S, Nill C, Zourelidou M, Dohmann EM, Maier A, Schwechheimer C (2007) The DELLA domain of GA INSENSITIVE mediates the interaction with the GA INSENSITIVE DWARF1A gibberellin receptor of Arabidopsis. Plant Cell 19(4):1209–1220. https://doi.org/10.1105/tpc.107.051441
Yang C, Lu X, Ma B, Chen SY, Zhang JS (2015) Ethylene signaling in rice and Arabidopsis: conserved and diverged aspects. Mol Plant 8(4):495–505. https://doi.org/10.1016/j.molp.2015.01.003
Acknowledgments
This project was supported by grants from Energy Biosciences Institute, US DOE DE-SC0010686, the International Consortium for Sugarcane Biotechnology, and startup fund from Fujian Agriculture and Forestry University to RM.
Author information
Authors and Affiliations
Contributions
RM and FZ conceived the study, RM coordinated all research activities, TJ and CN conducted field trials, FZ, CMW, and JZ carried out RNAseq experiment and bioinformatics analysis. FZ and RM wrote the manuscript; all authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Zhu, F., Wai, C.M., Zhang, J. et al. Differential expression of hormone related genes between extreme segregants of a Saccharum interspecific F2 population. Euphytica 214, 55 (2018). https://doi.org/10.1007/s10681-018-2137-z
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-018-2137-z