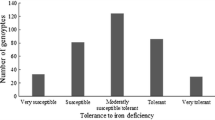
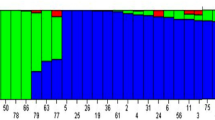
Abstract
Cowpea (Vigna unguiculata (L.) Walp) is a legume of economic importance world-wide, especially in Western Africa, where it is an important part of the population’s diet. The rapidly increasing population growth in Africa requires substantial increase in cowpea production, which can be achieved by expanding land areas for agricultural purposes. In addition, prevalence of soil acidity in Africa constrains such an alternative since phosphorus availability, a key element for plant growth and development, is limited, thus resulting in poor cowpea production. The objectives of this study were to conduct an association analysis for adaptation to low phosphorus conditions and rock phosphate response in cowpea, and to identify SNP markers associated with these two traits. A total of 357 cowpea accessions, collected worldwide, was evaluated for phosphorus stress and response to addition of rock phosphate. Association analysis was conducted using 1018 SNPs obtained using genotyping-by-sequencing (GBS). TASSEL 5 and R were used for association mapping studies based on six different models. The results indicated that: (1) substantial variability in adaptation to low phosphorus conditions and rock phosphate response exists in the USDA cowpea accession panel; (2) ten SNP markers, C35006753_110, C35028233_482, C35072764_1384, C35084634_455, Scaffold21750_4938, Scaffold26894_5408, Scaffold41885_14420, Scaffold45170_4650, Scaffold50732_679; and Scaffold88448_741 were found to be associated with tolerance to low phosphorus conditions in cowpea, and (3) eight SNP markers, C35028233_482, C35058535_121, Scaffold26894_5408, Scaffold45170_4650, Scaffold51609_507, Scaffold53730_7339, Scaffold74389_5733, and Scaffold87916_4921 were highly associated with rock phosphate response. These SNP markers can be used in a marker-assisted breeding (MAS) program to improve cowpea tolerance to phosphorus stress.



Similar content being viewed by others
References
Abaidoo R, Dare MO, Killani S (2017) Evaluation of early maturing cowpea (Vigna unguiculata) germplasm for variation in phosphorus use efficiency and biological nitrogen fixation potential. J Agric Sci 155(1):102–116
Agbicodo EM, Fatokun CA, Bandyopadhyay R, Wydra K, Diop NN, Muchero W, Ehlers JD, Roberts PA, Close TJ, Visser RGF, Van der Linden (2010) Identification of markers associated with bacterial blight resistance loci in cowpea [Vign aunguiculata (L.) Walp]. Euphytica 175:215–226
Araújo AP, Antunes IF, Teixeira MG (2005) Inheritance of root traits and phosphorus uptake in common bean (Phaseolus vulgaris L.) under limited soil phosphorus supply. Euphytica 145(1–2):33–40
Badiane FA, Diouf M, Diouf D (2014) Cowpea in broadening the genetic base of grain legumes. Springer, India, pp 95–114
Bastien M, Sonah H, Belzile F (2014) Genome wide association mapping of resistance in soybean with a genotyping-by-sequencing approach. Plant Genome. doi:10.3835/plantgenome2013.10.0030
Beebe SE, Rojas-Pierce M, Yan X, Matthew WB, Pedraza F, Muñoz F, Tohme J, Jonathan PL (2006) Quantitative trait loci for root architecture traits correlated with phosphorus acquisition in common bean. Crop Sci 46(1):413–423
Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES (2007) TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics 23(19):2633–2635
Caygill JC, Janis AJ, Carol EMF (1981) Imitation milks from Cicer arietinum (L.), Vigna unguiculata (L.) Walpers and Vigna radiata (L.) Wilczek and other legumes. J Sci Food Agric 32(6):601–607
Collard BCY, Mackill DJ (2008) Marker-assisted selection: an approach for precision plant breeding in the twenty-first century. Biol Sci 363(1491):557–572
Coltman RGG, Gabelman W (1982) Intraspecific variation in growth, phosphorus acquisition and phosphorus utilization in tomatoes under phosphorus-deficiency stress. In: Proceedings of the Ninth International Plant Nutrition Colloquium, Warwick University, England
Earl DA, VonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES, Mitchell SE (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS ONE 6(5):e19379
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14(8):2611–2620
Fawole I, Gabelman WH, Gerloff GC (1982) Heritability of efficiency in phosphorus utilization in beans (Phaseolus vulgaris L.) grown under phosphorus stress. J Am Soc Hortic Sci 107:94–97
Fita A, Nuez F, Picó B (2011) Diversity in root architecture and response to P deficiency in seedlings of Cucumis melo L. Euphytica 181(3):323–339
Ghaderi A, Lower RL (1979) Gene effects of some vegetative characters of cucumber. J Am Soc Hortic Sci 104:141–144
Hall AE, Cisse N, Thiaw S, Elawad HO, Ehlers JD, Ismail AM, Fery RL, Roberts PA, Kitch LW, Murdock LL, Boukar O (2003) Development of cowpea cultivars and germplasm by the bean/cowpea CRSP. Field Crops Res 82(2):103–134
Hammond JP, Broadley MR, White PJ, King GJ, Bowen HC, Hayden R, Meacham MC, Mead A, Overs T, Spracklen WP, Greenwood DJ (2009) Shoot yield drives phosphorus use efficiency in Brassica oleracea and correlates with root architecture traits. J Exp Bot 60(7):1953–1968
Hayman DS (1975) Phosphorus cycling by soil microorganisms and plant roots. In: Walker N (ed) Soil microbiology. Butterworths, London, pp 67–92
Jones C, Hatier JH, Cao M, Fraser K, Rasmussen S (2015) Metabolomics of plant phosphorus-starvation response. Annu Plant Rev 48:215–236
Kisha TJ, Sneller CH, Diers BW (1997) Relationship between genetic distance among parents and genetic variance in populations of soybean. Crop Sci 37(4):1317–1325
Kumar S, Stecher G, Tamura K (2016) MEGA7: Molecular evolutionary genetics analysis Version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121(1):185–199
Li YD, Wang YJ, Tong YP, Gao JG, Zhang JS, Chen SY (2005) QTL mapping of phosphorus deficiency tolerance in soybean (Glycine max L. Merr.). Euphytica 142(1–2):137–142
Li R, Yu C, Li Y, Lam TW, Yiu SM, Kristiansen K, Wang J (2009) SOAP2: An improved ultrafast tool for short read alignment. Bioinformatics 25(15):1966–1967
Liang Q, Cheng X, Mei M, Yan X, Liao H (2010) QTL analysis of root traits as related to phosphorus efficiency in soybean. Ann Bot 106(1):223–234
Liu X, Huang M, Fan B, Buckler ES, Zhang Z (2016) Iterative usage of fixed and random effect models for powerful and efficient genome-wide association studies. PLoS Genet 12(2):e1005767
Mahamane S, Payne WA, Loeppert RH, Miller JC Jr, Reed DW (2008) Evaluation of cowpea genotypes for adaptation to low soil P conditions and rock phosphate application. HortScience 43(3):618–619
Mc Vickar MH, Bridger GL, Nelson LB (1963) Advances in phosphate fertilizers. Soil Sci Soc Am 17:155–187
Moose SP, Mumm RH (2008) Molecular plant breeding as the foundation for 21st century crop improvement. Plant Physiol 147(3):969–977
Ochoa IE, Blair MW, Lynch JP (2006) QTL analysis of adventitious root formation in common bean under contrasting phosphorus availability. Crop Sci 46(4):1609–1621
Olufajo OO (2012) Agronomic performance of improved cowpea varieties under natural infestation with Alectra vogelii (Benth.) in the northern Guinea savannah of Nigeria. Agric Trop Subtrop 45(2):66–71
Parentoni SN, de Souza Jr CL, de Carvalho Alves VM, Gama EE, Coelho AM, De Oliveira AC, Guimarães PE, Guimarães CT, Vasconcelos MJ, Patto Pacheco CA, Meirelles WF (2010) Inheritance and breeding strategies for phosphorous efficiency in tropical maize (Zea mays L.). Maydica 55(1):1–15
Payne WA, Wendt CW, Lascano RJ (1990) Root zone water balances of three low-input millet fields in Niger. West Afr Agron J 82(4):813–819
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–959
Qin J, Shi A, Xiong H, Mou B, Motes D, Lu W, Miller JC Jr, Scheuring DC, Nzaramba MN, Weng Y, Yang W (2016) Population structure analysis and association mapping of seed antioxidant content in USDA Cowpea (Vigna unguiculata L. Walp.) core collection using SNPs. Can J Plant Sci 96(6):1026–1036
Ramasamy RK, Ramasamy S, Bindroo BB, Naik VG (2014) STRUCTURE PLOT: a program for drawing elegant STRUCTURE bar plots in user friendly interface. SpringerPlus 3(1):431
Rothe JC (2014) Breeding for tolerance of cowpea to low phosphorus soil conditions through physiological and genetic studies. Ph.D. Dissertation, Texas A&M University
Shi A, Buckley B, Mou B, Motes D, Morris JB, Ma J, Xiong H, Qin J, Yang W, Chitwood J, Weng Y (2016) Association analysis of cowpea bacterial blight resistance in USDA cowpea germplasm. Euphytica 208(1):143–155
Thakur D, Kaushal R, Shyam V (2014) Phosphate solubilising microorganisms: role in phosphorus nutrition of crop plants-A review. Agric Rev 35(3):159–171
Varshney RK, Nayak SN, May GD, Jackson SA (2009) Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol 27(9):522–530
Veneklaas EJ, Lambers H, Bragg J, Finnegan PM, Lovelock CE, Plaxton WC, Price CA, Scheible WR, Shane MW, White PJ, Raven JA (2012) Opportunities for improving phosphorus-use efficiency in crop plants. New Phytol 195(2):306–320
Verdcourt B (1970) Studies in the Leguminosae-Papilionoïdeae for the flora of tropical East Africa. Kew Bull 24(3):507–569
Wang L, Butterly CR, Yang XL, Wang Y, Herath HM, Jiang X (2012) Use of crop residues with alkaline slag to ameliorate soil acidity in an ultisol. Soil Use Manag 28(2):148–156
Wissuwa M, Wegner J, Ae N, Yano M (2002) Substitution mapping of Pup1: a major QTL increasing phosphorus uptake of rice from a phosphorus-deficient soil. Theor Appl Genet 105(6–7):890–897
Xiong H, Shi A, Mou B, Qin J, Motes D, Lu W, Ma J, Weng Y, Yang W, Wu D (2016) Genetic diversity and population structure of cowpea (Vigna Unguiculata L. Walp). PLoS ONE 11(8):e0160941
Xu Y, Crouch JH (2008) Marker-assisted selection in plant breeding: from publications to practice. Crop Sci 48(2):391–407
Yan X, Liao H, Beebe SE, Blair MW, Lynch JP (2004) QTL mapping of root hair and acid exudation traits and their relationship to phosphorus uptake in common bean. Plant Soil 265(1–2):17–29
Zhang D, Zhang H, Chu S, Li H, Chi Y, Triebwasser-Freese D, Lv H, Yu D (2017) Integrating QTL mapping and transcriptomics identifies candidate genes underlying QTLs associated with soybean tolerance to low-phosphorus stress. Plant Mol Biol 93(1–2):137–150
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ravelombola, W., Qin, J., Shi, A. et al. Association mapping revealed SNP markers for adaptation to low phosphorus conditions and rock phosphate response in USDA cowpea (Vigna unguiculata (L.) Walp.) germplasm. Euphytica 213, 183 (2017). https://doi.org/10.1007/s10681-017-1971-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1971-8