Abstract
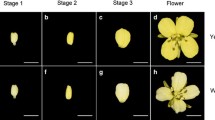
The petal color of B. napus is usually yellow, but white, cream, golden yellow, orange, and other colors have also been reported. The pigmentation mechanisms and evolutionary processes of petal color in B. napus remain elusive. An analysis of the inheritance of the orange petal color (OPC) trait indicated that this trait was controlled by two recessive genes, termed Bnpc1 and Bnpc2. For fine mapping of the Bnpc1 gene, two backcrossing populations, BC4 and BC5, were developed from the cross between an OPC parent, X118, and a yellow petal color parent, R258. Whole-genome re-sequencing with bulked segregation analysis using BC4 individuals was conducted, and as a result, two major candidate intervals of Bnpc1 were identified on chromosome C09 of B. napus (C09: 1.000–5.464 and 6.491–7.777 Mb). Subsequently, the genetic and physical linkage maps were constructed for the BC5 population using 10 simple sequence repeat, two sequence-characterized amplified region and 12 insertion-deletion (InDel) markers. The Bnpc1 gene was located in scaffold_38 on C09 of B. napus and flanked by Indel-31 and Indel-5, with genetic distances of 0.083 and 0.042 cM, respectively. The interval distance between the two markers was 151 kb (4.624–4.775 Mb) and was included in the first re-sequenced candidate region (C09: 1.000–5.464 Mb). The present study may facilitate cloning of the Bnpc1 gene as well as marker-assisted selection for developing OPC B. napus varieties and revealing the mechanism of petal color formation in B. napus.



Similar content being viewed by others
Abbreviations
- OPC :
-
Orange petal color
- YPC :
-
Yellow petal color
- ZEP :
-
Zeaxanthin epoxidase
- InDel:
-
Insertion-deletion
- SSR:
-
Simple sequence repeat
- SCAR:
-
Sequence-characterized amplified region
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M, Innan H, Cano L, Kamoun S, Terauchi R (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174–178. doi:10.1038/nbt.2095
Auldridge ME, Block A, Vogel JT, Dabney-Smith C, Mila I, Bouzayen M, Magallanes-Lundback M, DellaPenna D, McCarty DR, Klee HJ (2006) Characterization of three members of the A. thaliana carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family. Plant J 45:982–993. doi:10.1111/j.1365-313X.2006.02666.x
Brown BA, Clegg MT (1984) Influence of flower color polymorphism on genetic transmission in a natural population of the common morning glory, Ipomoea purpurea. Evolution 38:796–803. doi:10.1111/j.1558-5646.1984.tb00352.x
Caruso CM, Scott SL, Wray JC, Walsh CA (2010) Pollinators, herbivores, and the maintenance of flower color variation: a case study with Lobelia siphilitica. Int J Plant Sci 171:1020–1028. doi:10.1086/656511
Chalhoub B, Denoeud F, Liu S, Parkin IAP, Tang H, Wang X, Chiquet J, Belcram H, Tong C, Samans B, Corréa M, Da Silva C, Just J, Falentin C, Koh CS, Le Clainche I, Bernard M, Bento P, Noel B, Labadie K, Alberti A, Charles M, Arnaud D, Guo H, Daviaud C, Alamery S, Jabbari K, Zhao M, Edger PP, Chelaifa H, Tack D, Lassalle G, Mestiri I, Schnel N, Le Paslier M-C, Fan G, Renault V, Bayer PE, Golicz AA, Manoli S, Lee T-H, Thi VHD, Chalabi S, Hu Q, Fan C, Tollenaere R, Lu Y, Battail C, Shen J, Sidebottom CHD, Wang X, Canaguier A, Chauveau A, Bérard A, Deniot G, Guan M, Liu Z, Sun F, Lim YP, Lyons E, Town CD, Bancroft I, Wang X, Meng J, Ma J, Pires JC, King GJ, Brunel D, Delourme R, Renard M, Aury J-M, Adams KL, Batley J, Snowdon RJ, Tost J, Edwards D, Zhou Y, Hua W, Sharpe AG, Paterson AH, Guan C, Wincker P (2014) Early allopolyploid evolution in the post-Neolithic Brassica napus oilseed genome. Science 345:950. doi:10.1126/science.1253435
Cheng X, Xu J, Xia S, Gu J, Yang Y, Fu J, Qian X, Zhang S, Wu J, Liu K (2009) Development and genetic mapping of microsatellite markers from genome survey sequences in Brassica napus. Theor Appl Genet 118:1121–1131. doi:10.1007/s00122-009-0967-8
Chiou CY, Pan HA, Chuang YN, Yeh KW (2010) Differential expression of carotenoid-related genes determines diversified carotenoid coloration in floral tissues of Oncidium cultivars. Planta 232:937–948. doi:10.1007/s00425-010-1222-x
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Faegri K, Pijl LVD (1979) The principles of pollination ecology. Pergamon Press, New York
Feng FJ, Luo LJ, Li Y, Zhou LG, Xu XY, Wu JH, Chen HW, Chen L, Mei HW (2005) Comparative analysis of polymorphism of InDel and SSR markers in rice. Mol Plant Breed 21:243–255
Fenster CB, Armbruster WS, Wilson P, Dudash MR, Thomson JD (2004) Pollination syndromes and floral specialization. Annu Rev Ecol Evol Syst 35:375–403. doi:10.1146/annurev.ecolsys.34.011802.132347
Geleta M, Heneen WK, Stoute AI, Muttucumaru N, Scott RJ, King GJ, Kurup S, Bryngelsson T (2012) Assigning Brassica microsatellite markers to the nine C-genome chromosomes using Brassica rapa var. trilocularis-B. oleracea var. alboglabra monosomic alien addition lines. Theor Appl Genet 125:455–466. doi:10.1007/s00122-012-1845-3
Grant V (1993) Effects of hybridization and selection on floral isolation. Proc Natl Acad Sci USA 90:990–993. doi:10.1073/pnas.90.3.990
Grotewold E (2006) The genetics and biochemistry of floral pigments. Annu Rev Plant Biol 57:761–780. doi:10.1146/annurev.arplant.57.032905.105248
Han FQ, Yang C, Fang ZY, Yang LM, Zhuang M, Lv HH, Liu YM, Li ZS, Liu B, Yu HL, Liu XP, Zhang YY (2015) Inheritance and InDel markers closely linked to petal color gene (cpc-1) in Brassica oleracea. Mol Breed 35:160. doi:10.1007/s11032-015-0354-x
He J, Zhao X, Laroche A, Lu ZX, Liu H, Li Z (2014) Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci 5:484. doi:10.3389/fpls.2014.00484
Huang Z, Ban Y, Bao R, Zhang X, Xu A, Ding J (2014) Inheritance and gene mapping of the white flower in Brassica napus L. N Z J Crop Hortic Sci 42:111–117. doi:10.1080/01140671.2013.863211
Iniguez-Luy FL, Voort AV, Osborn TC (2008) Development of a set of public SSR markers derived from genomic sequence of a rapid cycling Brassica oleracea L. genotype. Theor Appl Genet 117:977–985. doi:10.1007/s00122-008-0837-9
Irwin RE, Strauss SY, Storz S, Emerson A, Guibert G (2003) The role of herbivores in the maintenance of a flower color polymorphism in wild radish. Ecology 84:1733–1743. doi:10.1890/0012-9658(2003)084[1733:TROHIT]2.0.CO;2
Jones KN, Reithel JS (2001) Pollinator-mediated selection on a flower color polymorphism in experimental populations of Antirrhinum (Scrophulariaceae). Am J Bot 88:447–454. doi:10.2307/2657109
Kato M, Tokumasu S (1976) The mechanism of increased seed fertility accompanied with the change of flower colour in Brassicoraphanus. Euphytica 25:761–767. doi:10.1007/BF00041616
Koes RE, Quattrocchio F, Mol JNM (1994) The flavonoid biosynthetic pathway in plants: function and evolution. BioEssays 16:123–132. doi:10.1002/bies.950160209
Kosambi DD (1943) The estimation of map distances from recombination values. Ann Eugen 12:172–175. doi:10.1111/j.1469-1809.1943.tb02321.x
Lee J, Izzah NK, Jayakodi M, Perumal S, Joh HJ, Lee HJ, Lee SC, Park JY, Yang KW, Nou IS, Seo J, Yoo J, Suh Y, Ahn K, Lee JH, Choi GJ, Yu Y, Kim H, Yang TJ (2015) Genome-wide SNP identification and QTL mapping for black rot resistance in cabbage. BMC Plant Biol 15:32. doi:10.1186/s12870-015-0424-6
Leshchiner I, Alexa K, Kelsey P, Adzhubei I, Austin-Tse CA, Cooney JD, Anderson H, King MJ, Stottmann RW, Garnaas MK, Ha S, Drummond IA, Paw BH, North TE, Beier DR, Goessling W, Sunyaev SR (2012) Mutation mapping and identification by whole-genome sequencing. Genome Res 22:1541–1548. doi:10.1101/gr.135541.111
Li H, Durbin R (2010) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25:1754–1760. doi:10.1093/bioinformatics/btp324
Li Q, Wan JM (2005) SSRHunter: development of a local searching software for SSR sites. Yi Chuan 27:808–810
Li M, Chen WJ, Yi DL (1999) Studies on the inheritance of CMS restorer R18 with red color flower in rapeseed (Brassica napus L.). Sci Agric Sin 32:27–30
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. doi:10.1093/bioinformatics/btp352
Liu HL (1985) Genetics and breeding in Rapeseeds. Science and Technology Press, Shanghai
Liu XP, Tu JX, Chen BY, Fu TD (2004) Identification of the linkage relationship between the flower colour and the content of erucic acid in the resynthesized Brassica napus L. Acta Genet Sin 31:357–362
Liu H, Du D, Guo S, Xiao L, Zhao Z, Zhao Z, Xing X, Tang G, Xu L, Fu Z, Yao Y, Duncan RW (2016) QTL analysis and the development of closely linked markers for days to flowering in spring oilseed rape (Brassica napus L.). Mol Breed 36:52. doi:10.1007/s11032-016-0477-8
Lu H, Lin T, Klein J, Wang S, Qi J, Zhou Q, Sun J, Zhang Z, Weng Y, Huang S (2014) QTL-seq identifies an early flowering QTL located near Flowering Locus T in cucumber. Theor Appl Genet 127:1491–1499. doi:10.1007/s00122-014-2313-z
McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA (2010) The genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res 20:1297–1303. doi:10.1101/gr.107524.110
Michelmore RW, Paran I, Kesseli RV (1991) Identification of markers linked to disease-resistance genes by bulked segregant analysis: a rapid method to detect markers in specific genomic regions by using segregating populations. Proc Natl Acad Sci USA 88:9828–9832. doi:10.1073/pnas.88.21.9828
Morice J (1960) Inheritance of flower colour in rape, B. napus L. var. oleifera Metzger. Ann Inst Nat Rech Agron Paris Ser B Ann Amelior Pl 10:155–168
Nielsen KM, Lewis DH, Morgan ER (2003) Characterization of carotenoid pigments and their biosynthesis in two yellow flowered lines of Sandersonia aurantiaca (Hook). Euphytica 130:25–34. doi:10.1023/A:1022328828688
Noda N, Aida R, Kishimoto S, Ishiguro K, Fukuchi-Mizutani M, Tanaka Y, Ohmiya A (2013) Genetic engineering of novel bluer-colored chrysanthemums produced by accumulation of delphinidin-based anthocyanins. Plant Cell Physiol 54:1684–1695. doi:10.1093/pcp/pct111
Ohmiya A, Kishimoto S, Aida R, Yoshioka S, Sumitomo K (2006) Carotenoid cleavage dioxygenase (CmCCD4a) contributes to white color formation in chrysanthemum petals. Plant Physiol 142:1193–1201. doi:10.1104/pp.106.087130
Parkin IA, Gulden SM, Sharpe AG, Lukens L, Trick M, Osborn TC, Lydiate DJ (2005) Segmental structure of the Brassica napus genome based on comparative analysis with Arabidopsis thaliana. Genetics 171:765–781. doi:10.1534/genetics.105.042093
Peng JC (2008) Acquirement of the milk-white flower rape mutant and genetic model of its progenies. J Anhui Agric Sci 36:1787–1788
Piquemal J, Cinquin E, Couton F, Rondeau C, Seignoret E, Doucet I, Perret D, Villeger MJ, Vincourt P, Blanchard P (2005) Construction of an oilseed rape (Brassica napus L.) genetic map with SSR markers. Theor Appl Genet 111:1514–1523. doi:10.1007/s00122-005-0080-6
Ramsay LD, Jennings DE, Kearsey MJ, Marshall DF, Bohuon EJ, Arthur AE, Lydiate DJ (1996) The construction of a substitution library of recombinant backcross lines in Brassica oleracea for the precision mapping of quantitative trait loci. Genome 39:558–567. doi:10.1139/g96-071
Sharma AB, Shinada T, Kifuji Y, Kitashiba H, Nishio T (2012) Molecular mapping of a male fertility restorer locus of Brassica oleracea using expressed sequence tag-based single nucleotide polymorphism markers and analysis of a syntenic region in Arabidopsis thaliana for identification of genes encoding pentatricopeptide repeat proteins. Mol Breed 30:1781–1792. doi:10.1007/s11032-012-9761-4
Simms EL, Bucher MA (1996) Pleiotropic effects of flower-color intensity on herbivore performance on Ipomoea purpurea. Evolution 50:957–963. doi:10.2307/2410871
Song Q, Johnson C, Wilson TE, Kumar A (2014) Pooled segregant sequencing reveals genetic determinants of yeast pseudohyphal growth. PLoS Genet 10:e1004570. doi:10.1371/journal.pgen.1004570
Takagi H, Abe A, Yoshida K, Kosugi S, Natsume S, Mitsuoka C, Uemura A, Utsushi H, Tamiru M, Takuno S, Innan H, Cano LM, Kamoun S, Terauchi R (2013) QTL-seq: rapid mapping of quantitative trait loci in rice by whole genome resequencing of DNA from two bulked populations. Plant J 74:174–183. doi:10.1111/tpj.12105
Takagi H, Tamiru M, Abe A, Yoshida K, Uemura A, Yaegashi H, Obara T, Oikawa K, Utsushi H, Kanzaki E, Mitsuoka C, Natsume S, Kosugi S, Kanzaki H, Matsumura H, Urasaki N, Kamoun S, Terauchi R (2015) MutMap accelerates breeding of a salt-tolerant rice cultivar. Nat Biotechnol 33:445–449. doi:10.1038/nbt.31881
Tian L, Niu Y, Yu Q, Guo S, Liu L (2009) Genetic analysis of white flower color with mixed model of major gene plus polygene in Brassica napus L. Sci Agric Sin 42:3987–3995
Treutter D (2005) Significance of flavonoids in plant resistance and enhancement of their biosynthesis. Plant Biol 7:581–591. doi:10.1055/s-2005-873009
Väli Ü, Brandström M, Johansson M, Ellegren H (2008) Insertion-deletion polymorphisms (indels) as genetic markers in natural populations. BMC Genet 9:8. doi:10.1186/1471-2156-9-8
Voorrips RE (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78
Wang Y, Xiao L, Guo S, An F, Du D (2016) Fine mapping and whole-genome resequencing identify the seed coat color gene in Brassica rapa. PLoS ONE 11:e0166464. doi:10.1371/journal.pone.0166464
Wen YC, Zhang SF, Wang JP, Zhu JC, Zhao L (2010) Genetic studies of white petals and selection of cytoplasmic male sterile line with white petals in Brassica napus L. Chin Agric Sci Bull 26:95–97
Woods DL, Séguin-Swartz G (1998) An investigation on linkage between white flower colour and erucic acid in summer rape. Can J Plant Sci 78:109–111. doi:10.4141/P96-151
Xiao L, Zhao Z, Du D, Yao Y, Xu L, Tang G (2012) Genetic characterization and fine mapping of a yellow-seeded gene in Dahuang (a Brassica rapa landrace). Theor Appl Genet 124:903–909. doi:10.1007/s00122-011-1754-x
Xiao L, Zhao H, Zhao Z, Du D, Xu L, Yao Y, Zhao Z, Xing X, Shang G, Zhao H (2013) Genetic and physical fine mapping of a multilocular gene Bjln1 in Brassica juncea to a 208-kb region. Mol Breed 32:373–383. doi:10.1007/s11032-013-9877-1
Yang H, Tao Y, Zheng Z, Li C, Sweetingham MW, Howieson JG (2012) Application of next-generation sequencing for rapid marker development in molecular plant breeding: a case study on anthracnose disease resistance in Lupinus angustifolius L. BMC Genomics 13:318. doi:10.1186/1471-2164-13-318
Yi B, Chen Y, Lei S, Tu J, Fu T (2006) Fine mapping of the recessive genic male-sterile gene (Bnms1) in Brassica napus L. Theor Appl Genet 113:643–650. doi:10.1007/s00122-006-0328-9
Yi ZH, Lu YF, Guo XQ, Hui MX, Zhang LG, Zhang MK (2012) Development of simple sequence repeat (SSR) and insertion/deletion (InDel) markers in Chinese cabbage (Brassica rapa ssp. pekinesis) and analysis of their transferability. J Agric Biotechnol 20:1398–1406
Zhang J, Pu H, Qi C, Fu S (2000) Inheritance of flower color character in oilseed rape (Brassica napus L.). Chin J Oil Crop Sci 22:1–4
Zhang B, Lu CM, Kakihara F, Kato M (2002) Effect of genome composition and cytoplasm on petal colour in resynthesized amphidiploids and sesquidiploids derived from crosses between Brassica rapa and Brassica oleracea. Plant Breed 121:297–300. doi:10.1046/j.1439-0523.2002.722295.x
Zhang B, Liu C, Wang Y, Yao X, Wang F, Wu J, King GJ, Liu K (2015) Disruption of a CAROTENOID CLEAVAGE DIOXYGENASE 4 gene converts flower colour from white to yellow in Brassica species. New Phytol 206:1513–1526. doi:10.1111/nph.13335
Zhao Z, Xiao L, Xu L, Xing X, Tang G, Du D (2017) Fine mapping the BjPl1 gene for purple leaf color in B2 of Brassica juncea L. through comparative mapping and whole-genome re-sequencing. Euphytica 213:80. doi:10.1007/s10681-017-1868-6
Acknowledgements
The authors are grateful to Dr. Kun Lu and Dr. Zhi Zhao for critical technical assistance.
Funding
This research was financially supported by the National Key Research and Development Plan of China (2016YFD0100202), the Applied Basic Research Program of Qinghai Province (2015-ZJ-704), the National System of Technology of the Rapeseed Industry (NYCYTX-00516), and the Key Laboratory of Spring Rapeseed Genetic Improvement (2017-ZJ-Y09).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
We do not have any commercial or associative conflicts of interest regarding the research in this paper.
Ethical approval
This article does not describe any studies involving human participants or animals.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Yao, Y., Li, K., Liu, H. et al. Whole-genome re-sequencing and fine mapping of an orange petal color gene (Bnpc1) in spring Brassica napus L. to a 151-kb region. Euphytica 213, 165 (2017). https://doi.org/10.1007/s10681-017-1959-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s10681-017-1959-4