Abstract
Sclerotinia stem rot, caused by the necrotrophic fungal pathogen Sclerotinia sclerotiorum, is a serious constraint to lettuce production worldwide. To understand the molecular mechanisms underlying the plant’s response to S. sclerotiorum infection, next-generation sequencing was used to analyze the transcriptional alterations in lettuce post-infection in a time course of a compatible interaction. A total of 28,485, 28,777 and 29,018 unigenes were obtained at 6 hours post-inoculation (hpi), 12 hpi and 24 hpi, respectively. Among these, 4104, 4316 and 4980 genes were up-regulated while 3977, 3818 and 3802 genes were down-regulated at 6 hpi, 12 hpi and 24 hpi, respectively. Functional classification of the differentially expressed genes (DEGs) indicated that 21, 24 and 30 metabolic pathways were affected at 6 hpi, 12 hpi and 24 hpi, respectively. Some pathways were related to the plant immune response, such as ‘oxidative phosphorylation’, ‘plant hormone signal transduction’ and the ‘MAPK signaling pathway’. Real time quantitative PCR (RT-qPCR) analysis of six selected DEGs further validated the RNA-seq results. In addition, some novel potential disease responsive genes, including superoxide dismutase, WRKY transcription factors, pectinesterase inhibitor and ethylene-responsive transcription factors were identified. Our results suggest that lettuce adopts multiple strategies in regulating plant immunity to S. sclerotiorum infection. Collectively, the study provides new insights in the interaction of the plant - S. sclerotiorum pathosystem and provides information for further characterization of genes involved in plant resistance against S. sclerotiorum.




Similar content being viewed by others
Data availability
All data that support the findings of this study are presented herein.
References
Adachi, H., Nakano, T., Miyagawa, N., Ishihama, N., Yoshioka, M., Katou, Y., Yaeno, T., Shirasu, K., & Yoshioka, H. (2015). WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in Nicotiana benthamiana. The Plant Cell, 27(9), 2645–2663.
Antico, C. J., Colon, C., Banks, T., & Ramonell, K. M. (2012). Insights into the role of jasmonic acid-mediated defenses against necrotrophic and biotrophic fungal pathogens. Frontiers in Biology, 7(1), 48–56.
Bashi, Z. D., Rimmer, S. R., Khachatourians, G. G., & Hegedus, D. D. (2013). Brassica napus polygalacturonase inhibitor proteins inhibit Sclerotinia sclerotiorum polygalacturonase enzymatic and necrotizing activities and delay symptoms in transgenic plants. Canadian Journal of Microbiology, 59(2), 79–86.
Benjamini, Y., & Yekutieli, D. (2001). The control of the false discovery rate in multiple testing under dependency. Annals of Statistics, 29(4), 1165–1188.
Bolton, M. D., Thomma, B. P., & Nelson, B. D. (2006). Sclerotinia sclerotiorum (lib.) de Bary: Biology and molecular traits of a cosmopolitan pathogen. Molecular Plant Pathology, 7, 1–16.
Chittem, K., Yajima, W. R., Goswami, R. S., & del Río Mendoza, L. E. (2020). Transcriptome analysis of the plant pathogen Sclerotinia sclerotiorum interaction with resistant and susceptible canola (Brassica napus) lines. PLoS One, 15(3), e0229844.
Clarkson, J. P., Phelps, K., Whipps, J. M., Young, C. S., Smith, J. A., & Watling, M. (2007). Forecasting Sclerotinia disease on lettuce: A predictive model for carpogenic germination of Sclerotinia sclerotiorum sclerotia. Phytopathology, 97, 621–631.
Donaldson, P. A., Anderson, T., Lane, B. G., Davidson, A. L., & Simmonds, D. H. (2001). Soybean plants expressing an active oligomeric oxalate oxidase from the wheat gf-2.8 (germin) gene are resistant to the oxalate-secreting pathogen Sclerotina sclerotiorum. Physiological and Molecular Plant Pathology, 59(6), 297–307.
Ellouze, O. E., Loukil, S., & Marzouki, M. N. (2011). Cloning and molecular characterization of a new fungal xylanase gene from Sclerotinia sclerotiorum S2. BMB Reports, 44, 653–658.
Funk, V. A., Susanna, A., Stuessy, T. F., & Robinson, H. (2009). Classification of Compositae, 41–52. International Association for Plant Taxonomy.
Gilbert, B. M., & Wolpert, T. J. (2013). Characterization of the LOV1-mediated, victorin-induced, cell-death response with virus-induced gene silencing. Molecular Plant-Microbe Interactions, 26, 903–917.
Grover, A. (2012). Plant chitinases: Genetic diversity and physiological roles. Critical Reviews in Plant Sciences, 31(1), 57–73.
Guimaraes, R. L., & Stotz, H. U. (2004). Oxalate production by Sclerotinia sclerotiorum deregulates guard cells during infection. Plant Physiology, 136, 3703–3711.
Guo, P., Li, Z., Huang, P., Li, B., Fang, S., Chu, J., & Guo, H. (2017). A tripartite amplification loop involving the transcription factor WRKY75, salicylic acid, and reactive oxygen species accelerates leaf senescence. The Plant Cell, 29(11), 2854–2870.
Jones, J. D., & Dangl, J. L. (2006). The plant immune system. Nature, 444(7117), 323–329.
Kabbage, M., Williams, B., & Dickman, M. B. (2013). Cell death control: The interplay of apoptosis and autophagy in the pathogenicity of Sclerotinia sclerotiorum. PLoS Pathogens, 9, e1003287.
Kawahara, Y., Oono, Y., Kanamori, H., Matsumoto, T., Itoh, T., & Minami, E. (2012). Simultaneous RNA-Seq analysis of a mixed transcriptome of rice and blast fungus interaction. PLoS One, 7, e49423.
Keinath, N. F., Kierszniowska, S., Lorek, J., Bourdais, G., Kessler, S. A., Shimosato-Asano, H., Grossniklaus, U., Schulze, W. X., Robatzek, S., & Panstruga, R. (2010). PAMP (pathogen associated molecular pattern)-induced changes in plasma membrane compartmentalization reveal novel components of plant immunity. Journal of Biological Chemistry, 285, 39140–39149.
Kim, D., Paggi, J. M., Park, C., Bennett, C., & Salzberg, S. L. (2019). Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nature Biotechnology, 37(8), 907–915.
Kim, K. S., Min, J. Y., & Dickman, M. B. (2008). Oxalic acid is an elicitor of plant programmed cell death during Sclerotinia sclerotiorum disease development. Molecular Plant-Microbe Interactions, 21, 605–612.
Liao, Y., Smyth, G. K., & Shi, W. (2014). featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics, 30(7), 923–930.
Liu, F., Li, X., Wang, M., Wen, J., Yi, B., Shen, J., Ma, C., Fu, T., & Tu, J. (2018). Interactions of WRKY 15 and WRKY 33 transcription factors and their roles in the resistance of oilseed rape to Sclerotinia infection. Plant Biotechnology Journal, 16(4), 911–925.
Liu, F., Wang, M., Wen, J., Yi, B., Shen, J., Ma, C., Tu, J., & Fu, T. (2015). Overexpression of barley oxalate oxidase gene induces partial leaf resistance to Sclerotinia sclerotiorum in transgenic oilseed rape. Plant Pathology, 64(6), 1407–1416.
Lorenzo, O., Piqueras, R., Sánchez-Serrano, J. J., & Solano, R. (2003). ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. The Plant Cell, 15(1), 165–178.
Love, M. I., Huber, W., & Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biology, 15(12), 1–21.
Ma, M., Tu, C., Luo, J., Lu, M., Zhang, S., & Xu, L. (2021). Metabolic and immunological effects of gut microbiota in leaf beetles at the local and systemic levels. Integrative Zoology., 16, 313–323.
Marzin, S., Hanemann, A., Sharma, S., Hensel, G., Kumlehn, J., Schweizer, G., & Röder, M. S. (2016). Are PECTINESTERASE INHIBITOR genes involved in mediating resistance to Rhynchosporium commune in barley? PLoS One, 11(3), e0150485.
Nuruzzaman, M., Sharoni, A. M., & Kikuchi, S. (2013). Roles of NAC transcription factors in the regulation of biotic and abiotic stress responses in plants. Frontiers in Microbiology, 4, 248.
Oliveira, M. B., de Andrade, R. V., Grossi-de-Sa, M. F., & Petrofeza, S. (2015). Analysis of genes that are differentially expressed during the Sclerotinia sclerotiorum-phaseolus vulgaris interaction. Frontiers in Microbiology, 6, 1–14.
Pusztahelyi, T. (2018). Chitin and chitin-related compounds in plant-fungal interactions. Mycology, 9(3), 189–201.
Ranjan, A., Jayaraman, D., Grau, C., Hill, J. H., Whitham, S. A., Ane, J.-M., Smith, D. L., et al. (2018). The pathogenic development of Sclerotinia sclerotiorum in soybean requires specific host NADPH oxidases. Molecular Plant Pathology, 19, 700–714.
Ranjan, A., Westrick, N. M., Jain, S., Piotrowski, J. S., Ranjan, M., Kessens, R., Stiegman, L., Grau, C. R., Conley, S. P., Smith, D. L., & Kabbage, M. (2019). Resistance against Sclerotinia sclerotiorum in soybean involves a reprogramming of the phenylpropanoid pathway and up-regulation of antifungal activity targeting ergosterol biosynthesis. Plant Biotechnology Journal, 17(8), 1567–1581.
Seifbarghi, S., Borhan, M. H., Wei, Y., Coutu, C., Robinson, S. J., & Hegedus, D. D. (2017). Changes in the Sclerotinia sclerotiorum transcriptome during infection of Brassica napus. BMC Genomics, 18(1), 1–37.
Son, G. H., Wan, J., Kim, H. J., Nguyen, X. C., Chung, W. S., Hong, J. C., & Stacey, G. (2012). Ethylene-responsive element-binding factor 5, ERF5, is involved in chitin-induced innate immunity response. Molecular Plant-Microbe Interactions, 25(1), 48–60.
Tang, L., Yang, G., Ma, M., Liu, X., Li, B., Xie, J., Fu, Y., Chen, T., Yu, Y., Chen, W., Jiang, D., & Cheng, J. (2020). An effector of a necrotrophic fungal pathogen targets the calcium-sensing receptor in chloroplasts to inhibit host resistance. Molecular Plant Pathology, 21, 686–701.
Vidhyasekaran, P. (2015). Plant hormone signaling systems in plant innate immunity (Vol. 2, pp. 27–122). Springer.
Walz, A., Zingen-Sell, I., Loeffler, M., & Sauer, M. (2008). Expression of an oxalate oxidase gene in tomato and severity of disease caused by Botrytis cinerea and Sclerotinia sclerotiorum. Plant Pathology, 57, 453–458.
Wang, Z., Wan, L., Xin, Q., Chen, Y., Zhang, X., Dong, F., Hong, D., & Yang, G. (2018). Overexpression of OsPGIP2 confers Sclerotinia sclerotiorum resistance in Brassica napus through increased activation of defense mechanisms. Journal of Experimental Botany, 69(12), 3141–3155.
Wang, Z., Wan, L., Zhang, X., Xin, Q., Song, Y., Hong, D., et al. (2021). Interaction between Brassica napus polygalacturonase inhibition proteins and Sclerotinia sclerotiorum polygalacturonase: Implications for rapeseed resistance to fungal infection. Planta, 253(2), 1–18.
Wani, S. H., Anand, S., Singh, B., Bohra, A., & Joshi, R. (2021). WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Reports, 40(7), 1071–1085.
Wei, L., Jian, H., Lu, K., Filardo, F., Yin, N., Liu, L., Qu, C., Li, W., du, H., & Li, J. (2016). Genome-wide association analysis and differential expression analysis of resistance to Sclerotinia stem rot in Brassica napus. Plant Biotechnology Journal, 14, 1368–1380.
Wen, Z., Tan, R., Zhang, S., Collins, P. J., Yuan, J., Du, W., et al. (2018). Integrating GWAS and gene expression data for functional characterization of resistance to white mould in soya bean. Plant Biotechnology Journal, 16(11), 1825–1835.
Xing, Y., Chen, W. H., Jia, W., & Zhang, J. (2015). Mitogen-activated protein kinase kinase 5 (MKK5)-mediated signalling cascade regulates expression of iron superoxide dismutase gene in Arabidopsis under salinity stress. Journal of Experimental Botany, 66(19), 5971–5981.
Yan, H., Jia, H., Chen, X., Hao, L., An, H., & Guo, X. (2014). The cotton WRKY transcription factor GhWRKY17 functions in drought and salt stress in transgenic Nicotiana benthamiana through ABA signaling and the modulation of reactive oxygen species production. Plant and Cell Physiology, 55(12), 2060–2076.
Yang, G., Tang, L., Gong, Y., Xie, J., Fu, Y., Jiang, D., Li, G., Collinge, D. B., Chen, W., & Cheng, J. (2018). A cerato-platanin protein SsCP1 targets plant PR1 and contributes to virulence of Sclerotinia sclerotiorum. New Phytologist, 217, 739–755.
Yu, G., Wang, L. G., Han, Y., & He, Q. Y. (2012). clusterProfiler: An R package for comparing biological themes among gene clusters. OMICS, 16(5), 284–287.
Zhang, H., He, L., & Cai, L. (2018). Transcriptome sequencing: RNA-seq. In Computational systems biology (pp. 15–27). Humana Press.
Zhou, F., Zhang, X. L., Li, J. L., & Zhu, F. X. (2014). Dimethachlon resistance in Sclerotinia sclerotiorum in China. Plant Disease, 98, 1221–1226.
Zou, J., Li, W., Zhang, Y., Song, W., Jiang, H., Zhao, J., et al. (2021). Identification of glutathione transferase gene associated with partial resistance to Sclerotinia stem rot of soybean using genome-wide association and linkage mapping. Theoretical and Applied Genetics, 134(8), 2699–2709.
Acknowledgements
This work was supported by the China Agriculture Research System of MOF and MARA (CARS-23-G27), Postdoctoral Innovation Project of Hubei Province (CN) and Youth Innovation Project of Wuhan Academy of Agricultural Sciences (QNCX202102).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Additional Supporting Information may be found in the online version of the article at the publisher’s website.
Supplementary information
Supplementary Table S1
(XLSX 11.5 kb)
Supplementary Table S2
(XLSX 44.3 kb)
Supplementary Table S3
(XLSX 27.3 kb)
Supplementary Table S4
(XLSX 10.5 kb)
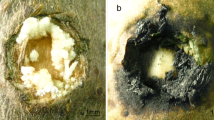
Fig. 5
Symptoms of S. sclerotiorum infection on lettuce leaves at 6, 12 and 24 hpi. Lettuce leaves were challenged with S. sclerotiorum strain 1980 and photographs (both the front side and the back side) were taken at each time point. (PNG 1.63 mb)
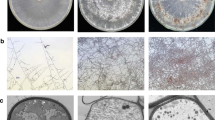
Fig. 6
Correlation between biological replicates. (a) Heat map of the correlation of gene expression levels from all samples compared with each other, represented by the value of Pearson’s correlation coefficient (R2). (b) Principal component analysis (PCA) of transcriptome expression. PCA plots of gene expression levels showing clustering of transcriptomes by time of sample collection. Ls0_1 / _2 / _3, Ls6_1 / _2 / _3, Ls12_1 / _2 /_3, Ls24_1 / _2 / _3: 0, 6, 12, 24 represent hours post inoculation, _1, _2, _3 represent three biological replicates. (PNG 1.14 mb)
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tang, L., Wang, B., Song, L. et al. RNAseq-based transcriptome analysis of lettuce infected by the necrotrophic fungus Sclerotinia Sclerotiorum. Eur J Plant Pathol 165, 85–96 (2023). https://doi.org/10.1007/s10658-022-02590-y
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-022-02590-y