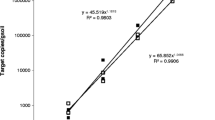
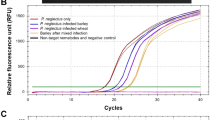
Abstract
PCR and qPCR are important methods for plant disease diagnosis and quantification of pathogen populations in host tissues or soil. For most plant diseases, DNA extracted from infected tissue or soil is a prerequisite for PCR or qPCR. The efficiency of DNA extraction should have a direct impact on the sensitivity of PCR and qPCR systems. In this study, plant pathogen DNA extraction efficiencies were evaluated based on three plant disease systems and three DNA extraction methods. DNA of bacterial (Pectobacterium atrosepticum), protist (Plasmodiophora brassicae) and fungal (Botrytis cinerea) pathogens was extracted and aliquots were prepared. Aliquots of the DNA were mixed with healthy tissues of the corresponding host plant and DNA was extracted again from the mixture. The resultant mixed DNA, as well as the original pathogen DNA, were analysed by qPCR to assess the quantities of the pathogen DNA. Extraction efficiency was calculated based on the qPCR Cq values of the mixed DNA and the original pathogen DNA for each extraction method on each pathogen. Our results indicated the pathogen DNA extraction efficiencies were generally less than, or near, 50%. This result called attention to the importance of: 1) including the DNA extraction efficiency in the evaluation and announcement of new qPCR diagnostic systems, and 2) developing and using high efficient DNA extraction methods in plant disease diagnosis and pathogen quantification.



Similar content being viewed by others
References
Abdullah, A. S., Turo, C., Moffat, C. S., Lopez-Ruiz, F. J., Gibberd, M. R., Hamblin, J., & Zerihun, A. (2018). Real-time PCR for diagnosing and quantifying co-infection by two globally distributed fungal pathogens of wheat. Frontiers in Plant Science, 9, 1086.
Al Rwahnih, M., Daubert, S., Golino, D., & Rowhani, A. (2009). Deep sequencing analysis of RNAs from a grapevine showing Syrah decline symptoms reveals a multiple virus infection that includes a novel virus. Virology, 387(2), 395–401.
Bahder, B. W., Soto, N., Komondy, L., Mou, D. F., Humphries, A. R., & Helmick, E. E. (2019). Detection and quantification of the 16SrIV-D phytoplasma in leaf tissue of common ornamental palm species in Florida using qPCR and dPCR. Plant Disease, 103(8), 1918–1922.
Buonicontro, D. S., Roberts, D. M., Oliveira, C. M. G., Blok, V. C., Neilson, R., & Oliveira, R. D. L. (2018). A rapid diagnostic for detection of Aphelenchoides besseyi and A. fujianensis based on real-time PCR. Plant Disease, 102(3), 519–526.
Bustin, S. A., Benes, V., Garson, J. A., Hellemans, J., Huggett, J., Kubista, M., Mueller, R., Nolan, T., Pfaffl, M. W., Shipley, G. L., Vandesompele, J., & Wittwer, C. T. (2009). The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clinical Chemistry, 55(4), 611–622.
Coetzee, B., Freeborough, M. J., Maree, H. J., Celton, J. M., Ress, D. J., & Burger, J. T. (2010). Deep-sequencing analysis of viruses infecting grapevines: Viome of a vineyard. Virology, 400(2), 157–163.
Dreo, T., Pirc, M., Ramšak, Ž., Pavšič, J., Milavec, M., Žel, J., & Gruden, K. (2014). Optimising droplet digital PCR analysis approaches for detection and quantification of bacteria: A case study of fire blight and potato brown rot. Analytical and Bioanalytical Chemistry, 406(26), 6513–6528.
Duan, Y. B., Ge, C. Y., Zhang, X. K., Wang, J. X., & Zhou, M. G. (2014). Development and evaluation of a novel and rapid detection assay for Botrytis cinerea based on loop-mediated isothermal amplification. PLoS One, 9(10), e111094.
Feng, J., Hwang, R., Chang, K. F., Hwang, S. B., Strelkov, S. E., Gossen, B. D., & Zhou, Q. (2010). An inexpensive method for extraction of genomic DNA from fungal mycelia. Canadian Journal of Plant Pathology, 32(3), 396-401.
Fu, H., Yang, Y., Mishra, V., Zhou, Q., Zuzak, K., Feindel, D., Harding, M. W., & Feng, J. (2020). Most Plasmodiophora brassicae populations in single canola root galls from Alberta fields are mixtures of multiple strains. Plant Disease, 104(1), 116–120.
Gallelli, A., Talocci, S., Pilotti, M., & Loreti, S. (2014). Real-time and qualitative PCR for detecting Pseudomonas syringae pv. actinidiae isolates causing recent outbreaks of kiwifruit bacterial canker. Plant Pathology, 63(2), 264–276.
Harper, S. J., Ward, L. I., & Clover, G. R. G. (2010). Development of a LAMP and real-time PCR methods for the rapid detection of Xylella fastidiosa for quarantine and field applications. Phytopathology, 100(2), 1282–1288.
Panno, S., Ruiz-Ruiz, S., Caruso, A. G., Alfaro-Fernandez, A., San Ambrosio, M. I. F., & Davino, S. (2019). Real-time reverse transcription polymerase chain reaction development for rapid detection of tomato brown rugose fruit virus and comparison with other techniques. Peer J, 7, e7928.
Rossmann, S., Dees, M. W., Perminow, J., Meadow, R., & Brurberg, M. B. (2018). Soft rot Enterobacteriaceae are carried by a large range of insect species in potato fields. Applied and Environmental Microbiology, 84(12), e00281–e00218.
Sarkes, A., Fu, H., Feindel, D., Harding, M., & Feng, J. (2020). Development and evaluation of a loop-mediated isothermal amplification (LAMP) assay for the detection of Tomato brown rugose fruit virus (ToBRFV). PLoS One, 15(6): e0230403.
Tomlinson, J. A., Barker, I., & Boonham, N. (2007). Faster, simpler, more-specific methods for improved molecular detection of Phytopthora ramorum in the field. Applied and Environmental Microbiology, 73(12), 4040–4047.
Wallenhammar, A. C., & Arwidsson, O. (2001). Detection of Plasmodiophora brassicae by PCR in naturally infested soils. European Journal of Plant Pathology, 107(3), 313–321.
Yang, Y., Zuzak, K., Harding, M., Strelkov, S., Hwang, S. B., Feindel, D., & Feng, J. (2018). DNA sequence dimorphisms in populations of the clubroot pathogen Plasmodiophora brassicae. Plant Disease, 102(9), 1703–1707.
Zahr, K., Sarkes, A., Yang, Y., Zhou, Q., Feindel, D., Harding, M., & Feng, J. (2019). Plasmodiophora brassicae in its environment-effects of temperature and light on resting spore survival in soil. bioRxiv. https://doi.org/10.1101/819524.
Funding
Financial support was received from Canadian Agricultural Partnership (no. 601322).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Research involving human and/or animals
No animals or data from human participants were involved in this study.
Supplementary Information
ESM 1
(PDF 41 kb)
Rights and permissions
About this article
Cite this article
Yang, Y., Zhou, Q., Zahr, K. et al. Impact of DNA extraction efficiency on the sensitivity of PCR-based plant disease diagnosis and pathogen quantification. Eur J Plant Pathol 159, 583–591 (2021). https://doi.org/10.1007/s10658-020-02189-1
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-020-02189-1