Abstract
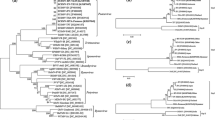
Bean common mosaic virus (BCMV) is an important plant pathogen causing considerable economic loss to legume production worldwide. In this study, the molecular characterizations of four BCMV isolates from Iran were determined, and their genetic variation and phylogenetic relationship compared with those of 49 other from GenBank. The genome contained a single open reading frame encoding a polyprotein of 3221, 3222, 3201 and 3202 amino acids (aa) in Ir-MSC1, Ir-GoB, Ir-MJB, and Ir-MSC2, respectively. Phylogenetic analysis of the polyprotein nucleotide sequences revealed that all 53 BCMV isolates clustered into six phylogroups distinguishable according to their genetic distances, namely S (soybean), P (peanut), C (common bean) and also three other distinct groups (R1, R2, and R3) of isolates all of which were recombinant. Iranian BCMV isolates did not form a compact group in the phylogenetic trees and shared 82.7–97.0% nt and 98.6–98.9% aa sequence identity among themselves and 80.1–98.7% nt and 83.4–99.8% aa sequence identity with the 49 other BCMV isolates. Of the 53 BCMV isolates studied, 34 (including Ir-MSC1 and Ir-GoB) were found to be recombinants at multiple regions of the genomes. The 19 non-recombinant isolates fell into three main phylogroups (S, P, and C) and tended to be classified according to their original host (with some exceptions). Statistically highly significant genetic differentiation (FST > 0.80) and infrequent gene flow were detected between the three phylogroups, suggesting ancestral differentiation. The normalized values for dN-dS indicated different purifying selection pressures acting on each of cistrons, with the P1 and NIa-Pro being subjected to the weakest and strongest functional constraints, respectively. Some codons (mainly in P1 and P3 regions) were under positive selection. This study presents a comprehensive analysis of evolutionary forces shaping the genetic structure of BCMV with implications for international exchanges of propagating materials.


Similar content being viewed by others
References
Adams, M. J., Antoniw, J. F., & Beaudoin, F. (2005). Overview and analysis of the polyprotein cleavage sites in the family Potyviridae. Molecular Plant Pathology, 6, 471–487.
Arli-Sokmen, M., Deligoz, I., & Kutluk-Yilmaz, N. D. (2016). Characterization of Bean common mosaic virus and Bean common mosaic necrosis virus isolates in common bean growing areas in Turkey. European Journal of Plant Pathology, 146, 1–16.
Berger, P. H., Wyatt, S. D., Shiel, P. J., Silbernagel, M. J., Druffel, K., & Mink, M. I. (1997). Phylogenetic analysis of the Potyviridae with emphasis on legume-infecting potyviruses. Archives of Virology, 142, 1979–1999.
Bitocchi, E., Nanni, L., Bellucci, E., Rossi, M., Giardini, A., Spagnoletti Zeuli, P., Logozzo, G., Stougaard, J., McClean, P., Attene, G. et al. (2012). Mesoamerican origin of the common bean (Phaseolus vulgaris L.) is revealed by sequence data. Proceedings of the National Academy of Sciences of the United States of America, Early Edition, 109(14), E788–796.
Bravo, E., Calverf, L. A., & Morales, F. J. (2008). The complete nucleotide sequence of the genomic RNA of Bean common mosaic virus strain NL4. Genetica, 32, 37–46.
Chare, E. R., & Holmes, E. C. (2006). A phylogenetic survey of recombination frequency in plant RNA viruses. Archives of Virology, 151, 933–946.
Chiquito-Almanza, E., Acosta-Gallegos, J. A., García-Álvarez, N. C., Garrido-Ramírez, E. R., Montero-Tavera, V., Guevara-Olvera, L., & Anaya-López, J. L. (2017). Simultaneous detection of both RNA and DNA viruses infecting dry bean and occurrence of mixed infections by BGYMV, BCMV and BCMNV in the central-west region of Mexico. Viruses, 9, e63.
Chowda-Reddy, R. V., Sun, H., Chen, H., Poysa, V., Ling, H., Gijzen, M., & Wang, A. (2011). Mutations in the P3 protein of Soybean mosaic virus G2 isolates determine virulence on Rsv4-genotype soybean. Molecular Plant-Microbe Interactions, 24, 37–43.
Chung, B. Y.-W., Miller, W. A., Atkins, J. F., & Firth, A. E. (2008). An overlapping essential gene in the Potyviridae. Proceedings of the National Academy of Sciences, USA, 105, 5897–5902.
Clark, M. F., & Adams, A. N. (1977). Characteristics of the micro plate method of enzyme-linked immunosorbent assay for the detection of plant viruses. Journal of General Virology, 34, 475–483.
Cronin, S., Verchot, J., Haldeman-Cahill, R., Schaad, M. C., & Carrington, J. C. (1995). Long-distance movement factor: A transport function of the potyvirus helper component proteinase. Plant Cell, 7, 549–559.
Daròs, J. A., & Carrington, J. C. (1997). RNA binding activity of NIa proteinase of tobacco etch potyvirus. Virology, 237, 327–336.
Daròs, J. A., Schaad, M. C., & Carrington, J. C. (1999). Functional analysis of the interaction between VPg-proteinase (NIa) and RNA polymerase (NIb) of tobacco etch potyvirus, using conditional and suppressor mutants. Journal of Virology, 73, 8732–8740.
Dolja, V. V., Haldeman, R., Robertson, N. L., Dougherty, W. G., & Carrington, J. C. (1994). Distinct functions of capsid protein in assembly and movement of tobacco etch potyvirus in plants. The EMBO Journal, 13, 1482–1491.
Drijfhout, E., & Morales, F. (2005). Bean common mosaic. In H. F. Schwartz, J. R. Steadman, R. Hall, & R. L. Forster (Eds.), Compendium of bean diseases (Secondth ed., pp. 60–62). St. Paul: American Phytopathological Society.
Drijfhout, E., Silbernagel, M. J., & Burke, D. W. (1978). Differentiation of strains of Bean common mosaic virus. Netherlands Journal of Plant Pathology, 84, 13–26.
Eggenberger, A., Hajimorad, M., & Hill, J. (2008). Gain of virulence on Rsv1-genotype soybean by an avirulent Soybean mosaic virus requires concurrent mutations in both P3 and HC-pro. Molecular Plant-Microbe Interactions, 21, 931–936.
Feng, X., Poplawsky, A. R., Nikolaeva, O. V., Myers, J. R., & Karasev, A. (2014a). Recombinants of Bean common mosaic virus (BCMV) and genetic determinants of BCMV involved in overcoming resistance in common bean. Phytopathology, 104, 786–793.
Feng, X., Poplawsky, A. R., & Karasev, A. V. (2014b). A recombinant of Bean common mosaic virus induces temperature-insensitive necrosis in an I gene-bearing line of common bean. Phytopathology, 104, 1251–1257.
Feng, X., Myers, J. R., & Karasev, A. (2015). Bean common mosaic virus isolate exhibits a novel pathogenicity profile in common bean, overcoming the bc-3 resistance allele coding for the mutated eIF4E translation initiation factor. Phytopathology, 105, 1487–1495.
Flores-Estevez, N., Acosta-Gallegos, J. A., & Silva-Rosales, L. (2003). Bean common mosaic virus and Bean common mosaic necrosis virus in Mexico. Plant Disease, 87, 21–25.
Fu, Y. X., & Li, W. H. (1993). Statistical tests of neutrality of mutations. Genetics, 133, 693–709.
García-Arenal, F., & McDonald, B. A. (2003). An analysis of the durability of resistance to plant viruses. Journal of Phytopathology, 93, 941–952.
García-Arenal, F., Fraile, A., & Malpica, J. M. (2001). Variability and genetic structure of plant virus populations. Annual Review of Phytopathology, 39, 157–186.
Gepts, P., & Bliss, F. A. (1988). Dissemination paths of common bean (Phaseolus vulgaris, Fabaceae) deduced from phaseolin electrophoretic variability. II. Europe and Africa. Economic Botany, 42, 86–104.
Gepts, P., Osborn, T. C., Rashka, K., & Bliss, F. A. (1986). Phaseolin protein variability in wild forms and landraces of the common bean (Phaseolus vulgaris)—Evidence for multiple centers of domestication. Economic Botany, 40, 451–468.
Gibbs, A. J., & Ohshima, K. (2010). Potyviruses and the digital revolution. Annual Review of Phytopathology, 48, 205–223.
Gibbs, A. J., Ohshima, K., Phillips, M. J., & Gibbs, M. J. (2008a). The prehistory of Potyviruses: Their initial radiation was during the dawn of agriculture. PLoS One, 3, e2523.
Gibbs, A. J., Trueman, J. W., & Gibbs, M. J. (2008b). The Bean common mosaic virus lineage of potyviruses: where did it arise and when? Archives of Virology, 153, 2177–2187.
Ha, C., Coombs, S., Revill, P. A., Harding, R. M., Vu, M., & Dale, J. L. (2008). Design and application of two novel degenerate primer pairs for the detection and complete genomic characterization of potyviruses. Archives of Virology, 153, 25–36.
Hampton, R. O., Silbernagel, M. J., & Burke, D. W. (1983). Bean common mosaic virus strains associated with bean mosaic epidemics in the northwestern United States. Plant Disease, 67, 658–661.
Hudson, R. R. (2000). A new statistic for detecting genetic differentiation. Genetics, 155, 2011–2014.
Huson, D. H., & Bryant, D. (2006). Application of phylogenetic networks in evolutionary studies. Molecular Biology and Evolution, 23, 254–267.
Johary, T., Dizadji, A., & Naderpour, M. (2016). Biological and molecular characteristics of Bean common mosaic virus isolates circulating in common bean in Iran. Journal of Plant Pathology, 98, 301–310.
Kim, B. M., Suehiro, N., Natsuaki, T., Inukai, T., & Masuta, C. (2010). The P3 protein of Turnip mosaic virus can alone induce hypersensitive response-like cell death in Arabidopsis thaliana carrying TuNI. Molecular Plant-Microbe Interactions, 23, 144–152.
Klein, R. E., Wyatt, S. D., & Kaiser, W. J. (1988). Incidence of Bean common mosaic virus in USDA Phaseolus germ plasm collection. Plant Disease, 72, 301–302.
Koenig, R., & Gepts, P. (1989). Allozyme diversity in wild Phaseolus vulgaris: Further evidence for two major centers of genetic diversity. Theoretical and Applied Genetics, 78, 809–817.
Larsen, R. C., Miklas, P. N., Druffel, K. L., & Wyatt, S. D. (2005). NL-3 K strain is a stable and naturally occurring interspecific recombinant derived from Bean common mosaic necrosis virus and Bean common mosaic virus. Phytopathology, 95, 1037–1042.
Larsen, R. C., Druffel, K. L., & Wyatt, S. D. (2011). The complete nucleotide sequences of Bean common mosaic necrosis virus strains NL-5, NL-8 and TN-1. Archives of Virology, 156, 729–732.
Li, Y. Q., Liu, Z. P., Yang, Y. S., Zhao, B., Fan, Z. F., & Wan, P. (2014). First report of Bean common mosaic virus infecting azuki bean (Vigna angularis) in China. Plant Disease, 98, 1017.
Li, Y., Cao, Y., & Wan, P. (2016). Identification of a naturally occurring Bean common mosaic virus recombinant isolate infecting azuki bean. Journal of Plant Pathology, 98, 129–133.
López, C., Aramburu, J., Galipienso, L., Soler, S., Nuez, F., & Rubio, L. (2011). Evolutionary analysis of tomato Sw-5 resistance-breaking isolates of Tomato spotted wilt virus. Journal of General Virology, 92, 210–215.
Maliogka, V. I., Salvador, B., Carbonell, A., Saenz, P., Leon, D. S., Oliveros, J. C., Delgadillo, M. O., Garcia, J. A., & Simon-mateo, C. (2012). Virus variants with differences in the P1 protein coexist in a Plum pox virus population and display particular host-dependent pathogenicity features. Molecular Plant Pathology, 13, 877–886.
Martin, D. P., Murrell, B., Golden, M., Khoosal, A., & Muhire, B. (2015). RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution, 1, vev003.
Martinez, F., Rodrigo, G., Aragones, V., Ruiz, M., Lodewijk, I., Fernandez, U., Elena, S. F., & Daros, J. A. (2016). Interaction network of tobacco etch potyvirus NIa protein with the host proteome during infection. BMC Genomics, 17, 87.
Mavric, I., & Sustar-Vozlic, J. (2004). Virus diseases and resistance to Bean common mosaic and Bean common mosaic necrosis potyvirus in common bean (Phaseolus vulgaris L.). Acta agriculturae slovenica, 83, 181–190.
Mink, G. I., & Silbernagel, M. J. (1992). Serological and biological relationships among viruses in the Bean common mosaic virus subgroup. Archives of Virology. Supplementum, 5, 397–406.
Moradi, Z., Mehrvar, M., Nazifi, E., & Zakiaghl, M. (2017). Iranian johnsongrass mosaic virus: the complete genome sequence, molecular and biological characterization, and comparison of coat protein gene sequences. Virus Genes, 53, 77–88.
Morales, F. J., & Bos, L. (1988). Bean common mosaic virus. Descriptions of Plant Viruses, No. 337. http://www.dpvweb.net/dpv/showdpv.php?dpvno=337.
Muhire, B. M., Varsani, A., & Martin, D. P. (2014). SDT: A virus classification tool based on pairwise sequence alignment and identity calculation. PLoS One, 9, e108277. https://doi.org/10.1371/journal.pone.0108277.
Murphy, J. F., Klein, P. G., Hunt, A. G., & Shaw, J. G. (1996). Replacement of the tyrosine residue that links a potyviral VPg to the viral RNA is lethal. Virology, 220, 535–538.
Naderpour, M., Mohammadi, M., Mossahebi, G. H., & Koohi Habibi, M. (2010). Identification of three strains of Bean common mosaic necrosis virus in common bean from Iran. Plant Disease, 94, 127.
Ogawa, T., Tomitaka, Y., Nakagawa, A., & Ohshima, K. (2008). Genetic structure of a population of Potato virus Y inducing potato tuber necrotic ringspot disease in Japan; comparison with north American and European populations. Virus Research, 131, 199–212.
Ohshima, K., Yamaguchi, Y., Hirota, R., Hamamoto, T., Tomimura, K., Tan, Z., Sano, T., Azuhata, F., Walsh, J. A., & Fletcher, J. (2002). Molecular evolution of Turnip mosaic virus: Evidence of host adaptation, genetic recombination and geographical spread. Journal of General Virology, 83, 1511–1521.
Olspert, A., Chung, B. Y., Atkins, J. F., Carr, J. P., Firth, A., & E. (2015). Transcriptional slippage in the positive-sense RNA virus family Potyviridae. EMBO Reports, e, 995–1004.
Parto, S., & Lartillot, N. (2018). Molecular adaptation in rubisco: Discriminating between convergent evolution and positive selection using mechanistic and classical codon models. PLoS One, 13, e0192697.
Pérez-Losada, M., Porter, M., & Crandall, K. A. (2008). Methods for analyzing viral evolution. In M. J. Roossinck (Ed.), Plant virus evolution (pp. 165–204). Berlin: Springer.
Pond, S. L. K., Frost, S. D. W., & Muse, S. V. (2005). HyPhy: Hypothesis testing using phylogenies. Bioinformatics, 21, 676–679.
Revers, F., Le Gall, O., Candresse, T., & Maule, A. J. (1999). New advances in understanding the molecular biology of plant: Potyvirus interactions. Molecular Plant-Microbe Interactions, 12, 367–376.
Riechmann, J. L., Lain, S., & Garcia, J. A. (1992). Highlights and prospects of potyvirus molecular biology. Journal of General Virology, 73, 1–16.
Rozas, J., Sanchez-DeI Barrio, J. C., Messeguer, X., & Rozas, R. (2003). DnaSP, DNA polymorphism analyses by the coalescent and other methods. Bioinformatics, 19, 2496–2497.
Rozas, J., Ferrer-Mata, A., Sánchez-DelBarrio, J. C., Guirao-Rico, S., Librado, P., Ramos-Onsins, S. E., & Sanchez-Gracia, A. (2017). DnaSP 6: DNA sequence polymorphism analysis of large datasets. Molecular Biology and Evolution, 34, 3299–3302.
Saqib, M., Nouri, S., Cayford, B., Jones, R. A. C., & Jones, M. G. K. (2010). Genome sequences and phylogenetic placement of two isolates of Bean common mosaic virus from Macroptilium atropurpureum in north-West Australia. Australasian Plant Pathology, 39, 184–191.
Seo, J. K., Ohshima, K., Lee, H. G., Son, M., Choi, H. S., Lee, S. H., Sohn, S. H., & Kim, K. H. (2009). Molecular variability and genetic structure of the population of Soybean mosaic virus based on the analysis of complete genome sequences. Virology, 393, 91–103.
Shahraeen, N., Ghotbi, T., Dizaje, A., & Sahandi, A. (2005). A survey of viruses affecting French bean (Phaseolus vulgaris L.) in Iran includes a first report of Southern bean mosaic virus and Bean pod mottle virus. Plant Disease, 89, 1012.
Sharma, P., Sharma, P. N., Kapil, R., Sharma, S. K., & Sharma, O. P. (2011). Analysis of 3′-terminal region of Bean common mosaic virus strains infecting common bean in India. Indian Journal of Virology, 22, 37–43.
Sharma, A., Sharma, V., Sharma, P., et al. (2016). Molecular characterization and genome organization of temperature insensitive necrosis inducing strain of BCMV infecting common bean in India. Australasian Plant Pathology, 45, 219–227.
Shukla, D. D., Jilka, J., Tosic, M., & Ford, R. E. (1989). A novel approach to the serology of potyviruses involving affinity-purified polyclonal antibodies directed towards virus specific N termini of coat proteins. Journal of General Virology, 70, 13–23.
Shukla, D. D., Lauricella, R., & Ward, C. W. (1992). Serology of potyviruses: current problems and some solutions. Archives of Virology. Supplementa, 5, 57–69.
Singh, S. P., & Schwartz, H. F. (2010). Breeding common bean for resistance to diseases. Review, 50, 2199–2223.
Skotnicki, M. L., Mackenzie, A. M., & Gibbs, A. J. (1996). Genetic variation in populations of kennedya yellow mosaic tymovirus. Advances in Virology, 141, 99–110.
Spence, N. J., & Walkey, D. G. A. (1995). Variation for pathogenicity among isolates of Bean common mosaic virus in Africa and reinterpretation of the genetic relationship between cultivars of Phaseolus vulgaris and pathotypes of BCMV. Plant Pathology, 44, 527–546.
Sztuba-Solińska, J., Urbanowicz, A., Figlerowicz, M., & Bujarski, J. J. (2011). RNA-RNA recombination in plant virus replication and evolution. Annual Review of Phytopathology, 49, 415–443.
Tajima, F. (1989). Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics, 123, 585–595.
Tamura, K., Stecher, G., Peterson, D., Filipski, A., & Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution, 30, 2725–2729.
Urcuqui-Inchima, S., Haenni, A., & Bernardi, F. (2001). Potyvirus proteins: a wealth of functions. Virus Research, 74, 157–175.
Valli, A., Jose, J., pez-Moya, L., & Garcia, J. A. (2007). Recombination and gene duplication in the evolutionary diversification of P1 proteins in the family Potyviridae. Journal of General Virology, 88, 1016–1028.
Van der Walt, E., Rybicki, E. P., Varsani, A., Polston, J. E., Billharz, R., Donaldson, L., Monjane, A. L., & Martin, D. P. (2009). Rapid host adaptation by extensive recombination. Journal of General Virology, 90, 734–746.
Verchot, J., Koonin, E. V., & Carrington, J. C. (1991). The 35-kDa protein from the N-terminus of the potyviral polyprotein functions as a third virus-encoded proteinase. Virology, 185, 527–535.
Verma, P., & Gupta, U. P. (2010). Immunological detection of Bean common mosaic virus in French bean (Phaseolusvulgaris L.) leaves. Indian Journal of Microbiology, 50, 263–265.
Vetten, H., Green, S., & Lesemann, D. E. (1992). Characterization of Peanut stripe virus isolates from soybean in Taiwan. Journal of Phytopathology, 135, 107–124.
Wright, S. (1951). The genetical structure of populations. Annals of Eugenics, 15, 323–354.
Zheng, H., Chen, J., Chen, J., Adams, M. J., & Hou, M. (2002). Bean common mosaic virus isolates causing different symptoms in asparagus bean in China differ greatly in the 5′-parts of their genomes. Archives of Virology, 147, 1257–1262.
Zhou, G.-C., Wu, X.-Y., Zhang, Y.-M., Wu, P., Wu, X.-Z., Liu, L.-W., Wang, Q., Hang, Y.-Y., Yang, J.-Y., Shao, Z.-Q., Wang, B., & Chen, J. Q. (2014). A genomic survey of thirty soybean-infecting Bean common mosaic virus (BCMV) isolates from China pointed BCMV as a potential threat to soybean production. Virus Research, 191, 125–133.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Electronic supplementary material
ESM 1
(PDF 417 kb)
Rights and permissions
About this article
Cite this article
Moradi, Z., Mehrvar, M. Genetic variability and molecular evolution of Bean common mosaic virus populations in Iran: comparison with the populations in the world. Eur J Plant Pathol 154, 673–690 (2019). https://doi.org/10.1007/s10658-019-01690-6
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-019-01690-6