Abstract
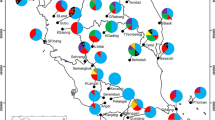
Aphanomyces euteiches Drechsler is an oomycete pathogen of leguminous crops that causes root rot, a severe disease of pea (Pisum sativum L.) worldwide. An improved understanding of the genetic structure of A. euteiches populations would increase knowledge of pathogen evolution and assist in the design of strategies to develop pea cultivars and germplasm with stable disease resistance. Twenty six primers pairs were used to amplify Sequence Related Amplified Polymorphisms (SRAP) among 49 A. euteiches isolates sampled from pea. A total of 190 polymorphic SRAP bands were generated, of which 82 were polymorphic between all the A. euteiches isolates. The percentage of polymorphic bands per primer pair ranged from 22 to 75%. According to the PIC value estimated for each marker, 60% of the SRAP markers were highly to reasonably informative (PIC > 0.25). Genetic structure of A. euteiches populations sampled in different American and French locations showed low to high genetic diversity within populations. The largest variation occurred within countries, with a total estimated genetic diversity of 0.477 and 0.172 for American and French populations, respectively. This was particularly evident from a principal component analysis (PCA) and a Minimum Spanning Networks (MSN) based on genetic profiles of isolates, which generated two different clusters, one corresponding to the French isolates and four American isolates (MV1, MV5, MV7, Ath3), and the other to American isolates. A. euteiches populations from cultivated pea in France appeared as a single unstructured population, whereas American isolates of A. euteiches diverged into three different populations.


Similar content being viewed by others
References
Botstein, D., White, R. L., Snolnick, M., & Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. American Journal of Human Genetics, 32, 314–331.
Burnett, V. F., Coventry, D. R., Hirth, J. R., & Greenhalgh, F. C. (1994). Subterranean clover decline in permanent pastures in north-eastern Victoria. Plant and Soil, 164, 231–241.
Burt, A., Carter, D. A., Koenig, G. L., White, T. J., & Taylor, J. W. (1996). Molecular markers reveal cryptic sex in the human pathogen Coccioides imitis. P Natl Acad Sci USA, 93.
Chen, C., Huang, L., Buchenauer, H., Zhao, H., Zuo, Y., & Kang, Z. (2009). Diversity among single zoospore isolates derived from single-zoo-sporangia of Phytophthora sojae Kauf. And Gerd. Journal of Phytopathology, 157, 181–187.
De Meeus, T., McCoy, K. D., Prugnolle, F., Chevillon, C., Durand, P., Hurtrez-Boussès, S., & Renaud, F. (2007). Population genetics and molecular epidemiology or how to “débusquer la bête”. Infection, Genetics and Evolution, 7, 308–332.
Didelot, D., Chaillet, I. (1995). Relevance and interest of root disease prediction tests for pea crop in France. In: Aep, ed. Proceedings of the 2nd European Conference on Grain Legumes – Improving production and utilisation of grain legumes, 1995. Copenhagen- Denmark, 150.
Excoffier, L., Smouse, P. E., & Quattro, J. M. (1992). Analysis of molecular variance inferred from metric distances among DNA haplotypes: Application to human mitochondrial DNA restriction data. Genetics, 131, 479–491.
Excoffier, L., Laval, G., & Schneider, S. (2005). Arlequin ver. 3.0: An integrated software package for population genetics data analysis. Evolutionary Bioinformatics Online, 1, 47–50.
Falush, D., Stephens, M., & Pritchard, J. K. (2003). Inference of population structure using multilocus genotype data: Linked loci and correlated allele frequencies. Genetics, 164, 1567–1587.
Gaulin, E., Jacquet, C., Bottin, A., & Dumas, B. (2007). Root rot disease of legumes caused by Aphanomyces euteiches. Molecular Plant Pathology, 8, 539–548.
Gaulin, E., Madoui, M. A., Bottin, A., Jacquet, C., Mathé, C., Couloux, A., Wincker, P., & Dumas, B. (2008). Transcriptome of Aphanomyces euteiches: New oomycete putative pathogenicity factors and metabolic pathways. PloS One, 3(3), e1723. doi:10.1371/journal.pone.0001723.
Goodwin, D. C., & Lee, S. B. (1993). Micorwave miniprep of total genomic DNA from fungi, plants, protists and animals for PCR. BioTechniques, 15, 438.
Greenhalgh, F. C., & Merriman, P. R. (1985). Aphanomyces euteiches, a cause of root rot of subterranean clover in Victoria. Australasian Plant Pathology, 14, 34–37.
Gritton. 1980. Field Pea. Hybridization of Crop Plants. p. 347–356. In: W.R. Fehr and H.H. Hadley (eds.), American Society of Agronomy, Inc., and Crop Science Society of America, Inc., Wisconsin.
Grünwald, N. J., & Hoheisel, G. A. (2006). Hierarchical analysis of diversity, selfing, and genetic differentiation in populations of the oomycete Aphanomyces euteiches. Phytopathology, 96, 1134–1141.
Grünwald, N. J., Goodwin, S. B., Milgroom, M. G., & Fry, W. E. (2003). Analysis of genotypic diversity data for populations of microorganisms. Phytopathology, 93, 738–746.
Hamon, C., Baranger, A., Coyne, C. J., McGee, R. J., Le Goff, I., L’Anthoëne, V., Esnault, R., Rivière, J.-P., Klein, A., Mangin, P., McPhee, K. E., Roux-Duparque, M., Porter, L., Miteul, H., Lesné, A., Morin, G., Onfroy, C., Moussart, A., Tivoli, B., Delourme, R., & Pilet-Nayel, M.-L. (2011). New consistent QTL in pea associated with partial resistance to Aphanomyces euteiches in controlled condition and multiple field environments from France and the United States of America. Theoretical and Applied Genetics, 123, 261–281.
Hamon, C., Coyne, C. J., McGee, R. J., Lesné, A., Esnault, R., Mangin, P., Hervé, M., Le Goff, I., Deniot, G., Roux-Duparque, M., Morin, G., McPhee, K. E., Delourme, R., Baranger, A., & Pilet-Nayel, M.-L. (2013). QTL meta-analysis provides a comprehensive view of the moderately low diversity of loci controlling partial resistance to Aphanomyces euteiches in four pea sources of resistance. BMC Plant Biology, 13, 45.
Holub, E. B., Grau, C. R., & Parke, J. L. (1991). Evaluation of the forma-specialis concept in Aphanomyces euteiches. Mycological Research, 95, 147–157.
Jombart, T. (2008). Adegenet: A R package for the multivariate analysis of genetic markers. Bioinformatics, 24, 1403–1405.
Jombart, T., Pontier, D., & Dufour, A. B. (2009). Genetic markers in the playground of multivariate analysis. Heredity, 102, 330–341.
Jones, F. R., & Drechsler, C. (1925). Root rot of peas in the United States caused by Aphanomyces euteiches. Journal of Agricultural Research, 30, 293–325.
Jones, F. R., & Linford, M. B. (1925). Pea disease survey in Wisconsin. Wis. Agr. Expt. Sta. Res. Bul., 64, 1–31.
Kamvar, Z. N., Larsen, M. M., Kanaskie, A. M., Hansen, E. M., & Grunwald, N. J. (2015). Spatial and temporal analysis of populations of the sudden oak death pathogen in Oregon forests. Phytopathology, 105, 982–989.
Kraft, J. M., & Pfleger, F. L. (2001). Compendium of pea diseases and pests. In J. M. Kraft & F. L. Pfleger (Eds.), The disease compendium series of the American Phytopathological Society. The American Phytopathological Society: St. Paul.
Kuk, A. Y., Li, X., & Xu, J. (2013). A fast collapsed data method for estimating haplotype frequencies from pooled genotype data with applications to the study of rare variants. Statistics in Medicine, 32(8), 1343–1360.
Labrousse, F. (1933). Notes de pathologie vegetale. Rev Pathol Vég Entomol Agric, 19, 71–84.
Levenfors, J. P., Wilkstrom, M., Persson, L., & Gerhardson, B. (2003). Pathogenicity of Aphanomyces spp. from different leguminous crops in Sweden. European Journal of Plant Pathology, 109, 535–543.
Levi, A., & Thomas, C. E. (2007). DNA markers from different linkage regions of watermelon genome useful in differentiating among closely related watermelon genotypes. Hortscience, 42, 210–214.
Li, G., & Quiros, F. C. (2001). Sequence-related amplified polymorphism (SRAP) a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in brassica. Theoretical and Applied Genetics, 103, 445–461.
Li, Q., Dong, S., Zhang, W., Lin, R., Wang, C., Qian, D., Lun, Z. R., Song, H. Q., & Zhu, X. Q. (2009). Sequence-related amplified polymorphism, an effective molecular approach for studying genetic variation in Fasciola spp. of human and animal health significance. Electrophoresis, 30, 403–409.
Liu, L., Zhu, X., Gong, Y., Song, X., Wang, T., Zhao, L., & Wang, L. (2007). Genetic giversity analysis of radish germplasm with RAPD, AFLP and SRAP markers. Acta Horticulturae, 760, 125–130.
Lou, Y., Lin, X., He, Q., Guo, X., Huang, L., & Fang, W. (2010). Analysis on genetic relationship of Puji-bamboo species by AFLP and SRAP. Mol Plant Breed, 8, 83–88.
Lynch, M., & Milligan, B. G. (1994). Analysis of population genetic structure with RAPD markers. Molecular Ecology, 3, 91–99.
Malvick, D. K., & Grau, C. R. (2001). Characteristics and frequency of Aphanomyces euteiches races 1 and 2 associated with alfalfa in the Midwestern United States. Plant Disease, 85, 740–744.
Malvick, D. K., & Percich, J. A. (1998). Genotypic and pathogenic diversity among pea-1 infecting strains of Aphanomyces euteiches from central and western United States. Phytopathology, 88, 915–921.
Malvick, D. K., Grau, C. R., & Percich, J. A. (1998). Characterization of Aphanomyces euteiches strains based on pathogenicity test and random amplified polymorphic DNA analyses. Mycological Research, 102, 465–475.
Malvick, D. K., Grünwald, N. J., & Dyer, A. T. (2009). Population structure, races, and host range of Aphanomyces euteiches from alfalfa production fields in the Central USA. European Journal of Plant Pathology, 123, 171–182.
Mieuzet, L., Quillévéré, A., Pilet, M.-L., & Le May, C. (2016). Development and characterization of microsatellite markers for the oomuceta Aphanomyces euteiches. Fungal Genetics and Biology, 91, 1–5.
Milgroom, M. G., & Peever, T. L. (2003). Population biology of plant pathogens: The synthesis of plant disease epidemiology and population genetics. Plant Disease, 89, 608–617.
Milgroom, M. G., Sotirovski, K., Spica, D., Davis, J. E., Brewer, M. T., Milec, M., & Cortesi, P. (2008). Clonal population structure of the chestnut blight fungus in expanding ranges in southeastern Europe. Molecular Ecology, 17, 4446–4458.
Moussart, A., Onfroy, C., Lesné, A., Esquibet, M., Grenier, E., & Tivoli, B. (2007). Host status and reaction of Medicago truncatula accessions to infection by three major pathogens of pea (Pisum sativum) and alfalfa (Medicago sativa). European Journal of Plant Pathology, 117, 57–69.
Moussart, A., Wicker, E., Le Delliou, B., Abelard, J.-M., Esnault, R., Lemarchand, E., Rouault, F., Le Guennou, F., Pilet-Nayel, M.-L., Baranger, A., Rouxel, F., & Tivoli, B. (2008). Spatial distribution of Aphanomyces euteiches inoculum in a naturally infested pea field. European Journal of Plant Pathology, 122, 452–458.
Mueller, U. G., & Wolfenbarger, L. L. (1999). AFLP genotyping and fingerprinting. Trends in Ecology & Evolution, 14, 389–394.
Nei, M. (1973). Analysis of gene diversity in subdivided populations. Proceedings of the Natl. Acad. Sci. USA 70, 3321–3323
Pasquali, M., Komjati, H., Lee, D., & Bayles, R. (2010). SRAP technique efficiently generates polymorphisms in Puccinia striiformis isolates. Journal of Phytopathology, 158, 708–711.
Peakall, R., & Smouse, P. E. (2012). GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research – Anupdate. Bioinformat, 28, 2537–2539.
Pfender, W. F., & Hagedorn, D. J. (1982). Aphanomyces euteiches f. Sp. phaseoli, a causal agent of bean root and hypocotyl rot. Phytopathology, 72, 306–310.
Pilet-Nayel, M. L., Muehlbauer, F. J., McGee, R. J., Kraft, J. M., Baranger, A., & Coyne, J. C. (2005). Consistent quantitative trait loci in pea for partial resistance to Aphanomyces euteiches isolates from the United States and France. Phytopathology, 95, 1287–1293.
Pritchard, J. K., Stephens, M., & Donnelly, P. (2000). Inference of population structure using multilocus genotype data. Genetics, 155, 945–959.
Robarts, D.W.H., Wolfe, A.D. 2014. Sequence-related amplified polymorphism (SRAP) markers: A potential resource for studies in plant molecular biology. Appl. Plant Sci 2(7), doi: 10.3732/apps.1400017.
Rosendahl, S. (2007). Genetic structure of Aphanomyces euteiches in Denmark studied by AFLP (amplified fragment length polymorphism). In M.-L. Pilet-Nayel, A. Moussart, A. Allee, & A. Baranger (Eds.), Proceedings of the the third international Aphanomyces workshop on legumes, 2007 (pp. 28–29). INRA-UNIP: Rennes.
Scott, W. W. (1961). A monograph of the genus Aphanomyces. Blacksburg: Virginia Agricultural Experiment Station Technical Bulletin.
Sherwood, R. T., & Hagedorn, D. J. (1958). Determining the common root rot potential of pea fields. Wisc Agr Exp Sta B, 531, 3–12.
Sun, S. J., Gao, W., Lin, S. Q., Zhu, J., Xie, B. G., & Lin, Z. B. (2006). Analysis of genetic diversity in Ganoderma population with a novel molecular marker SRAP. Appl Microbiol Biot, 72, 537–543.
Sundheim, L., Wiggen, K. 1972. Aphanomyces euteiches on peas in Norway. Isolation technique, physiologic races, and soil indexing. Norges Landbrukshogsk Meld 51, 17.
Tivoli, B., Baranger, A., Sivasithamparam, K., & Barbetti, M. J. (2006). Annual Medicago - from a model crop challenged by a spectrum of necrotrophic pathogens to a model plant to explore the nature of disease resistance. Annals of Botany, 98, 1117–1128.
Travadon, R., Sache, I., Dutech, C., Stachowiak, A., Marquer, B., & Bousset, L. (2011). Absence of isolation by distance patterns at the regional scale in the fungal plant pathogen Leptosphaeria maculans. Fungal Biology, 115, 649–659.
Wang, D., Wu, J., Tan, W., Pu, Z., & Yan, W. (2007). Genetic relationship of high starch sweet potato germplasm of Sichuan by SRAP. Southwest China J Agric Sci, 20, 506–509.
Wicker, E. (2001). Diversité des populations françaises d'Aphanomyces euteiches, agent de la pourriture racinaire du pois: variabilité pathogène et moléculaire. Rennes: ENSA Rennes, PhD.
Wicker, E., & Rouxel, F. (2001). Specific behaviour of French Aphanomyces euteiches Drechs. Populations for virulence and aggressiveness on pea, related to isolates from Europe, America and New Zealand. European Journal of Plant Pathology, 107, 919–929.
Wicker, E., Hulle, M., & Rouxel, F. (2001). Pathogenic characteristics of isolates of Aphanomyces euteiches from pea in France. Plant Pathology, 50, 433–442.
Yu, M., Ma, B., Luo, X., Zheng, L., Xu, X., & Yang, Z. (2008). Molecular diversity of Auricularia polytricha revealed by inter-simple sequence repeat and sequence-related amplified polymorphism markers. Current Microbiology, 56, 240–245.
Acknowledgements
The authors would like to thank Emmanuel Wicker (CIRAD, La Réunion), and Hélène Magalon (Laboratoire d’écologie marine, Université de la Réunion) for their valuable advice in the phylogenetic analyses. Financial support through grants of the Plant Health and the Environment Division of INRA, Terres Univia, AGROCAMPUS Ouest, and the US Department of Agriculture (USDA), is gratefully acknowledged. The authors would like to thank Dr. C. Coyne (USDA-ARS, Pullman) for providing soils from US.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The research presented in this manuscript does not implicate nor Human Participants and/or Animals.
CLM and GV were the principal investigator of this research project. They conducted the research experiments and contributed to the data analysis and to drafting the manuscript. CO contributed to the A. euteiches strains baiting, and to the drafting of the manuscript. MLP, AB, AM and DA contributed to the drafting of the manuscript. All authors read and approved the manuscript.
Rights and permissions
About this article
Cite this article
Le May, C., Onfroy, C., Moussart, A. et al. Genetic structure of Aphanomyces euteiches populations sampled from United States and France pea nurseries. Eur J Plant Pathol 150, 275–286 (2018). https://doi.org/10.1007/s10658-017-1274-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10658-017-1274-x