Abstract
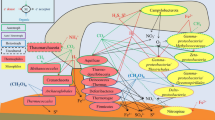
In this study, Illumina MiSeq sequencing of the 16 S rRNA gene was used to describe the bacterial communities in the South China Sea (SCS) during the southwest monsoon period. We targeted different regions in the SCS and showed that bacterial community was driven by the effects of the river, upwelling, and mesoscale eddy through changing the environmental factors (salinity, temperature, and nutrients). Distinct bacterial communities were observed among different chemical conditions, especially between the estuary and the open sea. The abundance of Burkholderiales, Frankiales, Flavobacteriales, and Rhodobacterales dominated the estuary and its adjacent waters. Bacteria in cyclonic eddy were dominated by Methylophilales and Pseudomonadales, whereas Prochlorococcus, SAR11 clade, and Oceanospirillales had relatively high abundance in the anticyclonic eddy. Overall, the abundance of specific phylotypes significantly varied among samples with different chemical conditions. Chemical conditions probably act as a driver that shapes and controls the diversity of bacteria in the SCS. This study suggests that the interaction between microbial and environmental conditions needs to be further considered to fully understand the diversity and function of marine microbes.





Similar content being viewed by others
References
Allen LZ, Allen EE, Badger JH, McCrow JP, Paulsen IT, Elbourne LDH, Thiagarajan M, Rusch DB, Nealson KH, Williamson SJ, Venter JC, Allen AE (2012) Influence of nutrients and currents on the genomic composition of microbes across an upwelling mosaic. ISME J 6:1403–1414
Allgaier M, Uphoff H, Felske A, Wagner-Dobler I (2003) Aerobic anoxygenic photosynthesis in Roseobacter clade bacteria from diverse marine habitats. Appl Environ Microbiol 69:5051–5059
Arrigo KR (2004) Marine microorganisms and global nutrient cycles. Nature 437:349–355
Azam F, Fenchel T, Field JG, Gray J, Meyer-Reil L, Thingstad F (1983) The ecological role of water-column microbes in the sea. Mar Ecol-Prog Ser 10:257–263
Azam F, Malfatti F (2007) Microbial structuring of marine ecosystems. Nat Rev Micro 5:782–791
Baltar F, Aristegui J, Gasol JM, Lekunberri I, Herndl GJ (2010) Mesoscale eddies: hotspots of prokaryotic activity and differential community structure in the ocean. ISME J 4:975–988
Benitez-Nelson CR, Bidigare RR, Dickey TD, Landry MR, Leonard CL, Brown SL, Nencioli F, Rii YM, Maiti K, Becker JW, Bibby TS, Black W, Cai WJ, Carlson CA, Chen FZ, Kuwahara VS, Mahaffey C, McAndrew PM, Quay PD, Rappe MS, Selph KE, Simmons MP, Yang EJ (2007) Mesoscale eddies drive increased silica export in the subtropical Pacific Ocean. Science 316:1017–1021
Blanchot J, André JM, Navarette C, Neveux J, Radenac MH (2001) Picophytoplankton in the equatorial Pacific: vertical distributions in the warm pool and in the high nutrient low chlorophyll conditions. Deep-Sea Res PT I: Oceanographic Research Papers 48:297–314
Brinkhoff T, Giebel H-A, Simon M (2008) Diversity, ecology, and genomics of the Roseobacter clade: a short overview. Arch Microbiol 189:531–539
Crump BC, Armbrust EV, Baross JA (1999) Phylogenetic analysis of particle-attached and free-living bacterial communities in the Columbia river, its estuary, and the adjacent coastal ocean. Appl Environl Microbiol 65:3192–3204
Crump BC, Hopkinson CS, Sogin ML, Hobbie JE (2004) Microbial biogeography along an estuarine salinity gradient: combined influences of bacterial growth and residence time. Appl Environ Microbiol 70:1494–1505
Eiler A, Hayakawa DH, Church MJ, Karl DM, Rappé MS (2009) Dynamics of the SAR11 bacterioplankton lineage in relation to environmental conditions in the oligotrophic North Pacific subtropical gyre. Environ Microbiol 11:2291–2300
Fuhrman JA (2009) Microbial community structure and its functional implications. Nature 459:193–199
Fuhrman JA, Hewson I, Schwalbach MS, Steele JA, Brown MV, Naeem S (2006) Annually reoccurring bacterial communities are predictable from ocean conditions. P Natl Acad Sci 103:13104–13109
Fuhrman JA, Steele JA, Hewson I, Schwalbach MS, Brown MV, Green JL, Brown JH (2008) A latitudinal diversity gradient in planktonic marine bacteria. P Natl Acad Sci 105:7774–7778
Galand PE, Potvin M, Casamayor EO, Lovejoy C (2010) Hydrography shapes bacterial biogeography of the deep Arctic Ocean. ISME J 4:564–576
Giovannoni SJ, Stingl U (2005) Molecular diversity and ecology of microbial plankton. Nature 437:343–348
Hamilton AK, Lovejoy C, Galand PE, Ingram RG (2008) Water masses and biogeography of picoeukaryote assemblages in a cold hydrographically complex system. Limnol Oceanogr 53:922–935
Hellerman S, Rosenstein M (1983) Normal monthly wind stress over the world ocean with error estimates. J Phys Oceanogr 13:1093–1104
Herlemann DPR, Labrenz M, Jurgens K, Bertilsson S, Waniek JJ, Andersson AF (2011) Transitions in bacterial communities along the 2000km salinity gradient of the Baltic Sea. ISME J 5:1571–1579
Hill PG, Zubkov MV, Purdie DA (2010) Differential responses of Prochlorococcus and SAR11-dominated bacterioplankton groups to atmospheric dust inputs in the tropical Northeast Atlantic Ocean. FEMS Microbiol Lett 306:82–89
Kalyuzhnaya MG, Bowerman S, Lara JC, Lidstrom ME, Chistoserdova L (2006) Methylotenera mobilis gen. nov., sp. nov., an obligately methylamine-utilizing bacterium within the family Methylophilaceae. Int J Syst Evol Micr 56:2819–2823
Lenk S, Moraru C, Hahnke S, Arnds J, Richter M, Kube M, Reinhardt R, Brinkhoff T, Harder J, Amann R, Musmann M (2012) Roseobacter clade bacteria are abundant in coastal sediments and encode a novel combination of sulfur oxidation genes. ISME J 6:2178–2187
Nan F, He Z, Zhou H, Wang D (2011) Three long-lived anticyclonic eddies in the northern South China Sea. J Geophys Res: Oceans 116:C05002
Nemergut DR, Costello EK, Hamady M, Lozupone C, Jiang L, Schmidt SK, Fierer N, Townsend AR, Cleveland CC, Stanish L, Knight R (2011) Global patterns in the biogeography of bacterial taxa. Environ Microbiol 13:135–144
Peterson TD, Crawford DW, Harrison PJ (2011) Evolution of the phytoplankton assemblage in a long-lived mesoscale eddy in the eastern Gulf of Alaska. Mar Ecol-Prog Ser 424:53–73
Pinhassi J, Gómez-Consarnau L, Alonso-Sáez L, Sala MM, Vidal M, Pedrós-Alió C, Gasol JM (2006) Seasonal changes in bacterioplankton nutrient limitation and their effects on bacterial community composition in the NW Mediterranean Sea. Aquat Microb Ecol 44:241–252
Radwan S, Al-Hasan R, Salamah S, Al-Dabbous S (2002) Bioremediation of oily sea water by bacteria immobilized in biofilms coating macroalgae. In biodeter biodegr 50:55–59
Sowell SM, Abraham PE, Shah M, Verberkmoes NC, Smith DP, Barofsky DF, Giovannoni SJ (2011) Environmental proteomics of microbial plankton in a highly productive coastal upwelling system. ISME J 5:856–865
Sun F-L, Wang Y-S, Wu M-L, Jiang Z-Y, Sun C-C, Cheng H (2014) Genetic diversity of bacterial communities and gene transfer agents in northern South China Sea. PLoS ONE 9:e111892
Sun F-L, Wang Y-S, Wu M-L, Jiang Z-Y, Sun C-C, Cheng H (2015) Spatial and vertical distribution of bacterial community in the northern South China Sea. Ecotoxicology 24:1478–1485
Sun F-L, Wang Y-S, Wu M-L, Wang Y-T, Li QP (2011) Spatial heterogeneity of bacterial community structure in the sediments of the Pearl River estuary. Biologia 66:574–584
Sun F-L, Wu M-L, Wang Y-S, Sun C-C, Xu Z-T (2020) Diversity and potential function of bacterial communities in different upwelling systems. Estuar, Coast Shelf S 237:106698
Xie S-P, Xie Q, Wang D, Liu WT (2003) Summer upwelling in the South China Sea and its role in regional climate variations. J Geophys Res: Oceans 108:3261
Zhang Y, Jiao N, Sun Z, Hu A, Zheng Q (2011) Phylogenetic diversity of bacterial communities in South China Sea mesoscale cyclonic eddy perturbations. Res Microbiol 162:320–329
Zhang Y, Sintes E, Chen J, Zhang Y, Dai M, Jiao N, Herndl GJ (2009) Role of mesoscale cyclonic eddies in the distribution and activity of archaea and bacteria in the South China Sea. Aquat Microb Ecol 56:65–79
Zhang Y, Zhao Z, Dai M, Jiao N, Herndl GJ (2014) Drivers shaping the diversity and biogeography of total and active bacterial communities in the South China Sea. Mol Ecol 23:2260–2274
Acknowledgements
This research was supported by the Project of Guangdong Science and Technology Department (2017A020216008), the National Natural Science Foundation of China (41406130, 31971480, U1901211 and 41876126), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA23050200 and XDA19060201), the Key Special Project for Introduced Talents Team of Southern Marine Science and Engineering Guangdong Laboratory (GML2019ZD0305), the National Key Research and Development Plan (2017FY100700) and the International Partnership Program of Chinese Academy of Sciences (133244KYSB20180012) and Innovation Academy of South China Sea Ecology and Environmental Engineering, Chinese Academy of Sciences (ISEE2019ZR02 and ISEE2018ZD02).
Funding
All the above funding has given the support.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Ethical approval
All the related authors confirmed there were no conflict of ethical approval.
Informed consent
All the related authors have known the informed consent.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
About this article
Cite this article
Sun, Fl., Wang, YS., Wu, ML. et al. Bacterial community variations in the South China Sea driven by different chemical conditions. Ecotoxicology 30, 1808–1815 (2021). https://doi.org/10.1007/s10646-021-02455-w
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10646-021-02455-w