Summary
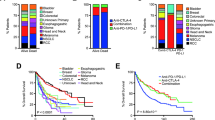
Introduction We analyzed the outcomes of patients with advanced cancers in our institution treated with off-label drugs targeting actionable genomic alteration based on next-generation sequencing who did not qualify for clinical trials. Purposes Our study endpoint was objective tumor response or stable disease at 16 weeks or later after treatment initiation. Methods Sixteen patients were included, 8 treated with immune checkpoint inhibitors targeting PD-L1 expression or TP53 mutations and 8 with other drugs. Tumors were analyzed based on PD-L1 expression, TP53 mutation, MSI, TMB, MMR status, and other targetable alterations. Results Of the 16 patients in the intention-to-treat group, no patients had an objective response after 16 weeks. Eleven patients met the primary study endpoint with stable disease, 8 in the immune checkpoint inhibitors group and 3 in the non-immune checkpoint inhibitors group. Using the log-rank test, the p-value for the difference between groups was 0.008. Conclusions In this study with off-label drugs, immune checkpoint inhibitors targeting TP53 mutations or PD-L1 expression were superior to the other drugs. This suggests the possibility of off-label use of anti-cancer drugs based on next-generation sequencing to be beneficial for advanced cancer patients without other therapeutic options.


Similar content being viewed by others
Data availability
We conducted an observational retrospective study. The datasets generated and analyzed were gathered from the EPIC database of the Mays Cancer Center, University of Texas Health MD Anderson Cancer Center and are available from the corresponding author upon reasonable request.
Code availability
Not applicable.
References
Mangat PK, Halabi S, Bruinooge SS, Garrett-Mayer E, Alva A, Janeway KA, Stella PJ, Voest E, Yost KJ, Perlmutter J, Pinto N, Kim ES, Schilsky RL (2018) Rationale and Design of the Targeted Agent and Profiling Utilization Registry (TAPUR) Study. JCO Precis Oncol 2018. https://doi.org/10.1200/PO.18.00122
Davis W, Makar G, Mehta P, Zhu GG, Somer R, Morrison J, Kubicek GJ (2019) Next-Generation Sequencing in 305 Consecutive Patients: Clinical Outcomes and Management Changes. J Oncol Pract 15(12):e1028–e1034. https://doi.org/10.1200/JOP.19.00269
Ott PA, Bang YJ, Piha-Paul SA, Razak ARA, Bennouna J, Soria JC, Rugo HS, Cohen RB, O'Neil BH, Mehnert JM, Lopez J, Doi T, van Brummelen EMJ, Cristescu R, Yang P, Emancipator K, Stein K, Ayers M, Joe AK, Lunceford JK (2019) T-Cell-Inflamed Gene-Expression Profile, Programmed Death Ligand 1 Expression, and Tumor Mutational Burden Predict Efficacy in Patients Treated With Pembrolizumab Across 20 Cancers: KEYNOTE-028. J Clin Oncol 37(4):318–327. https://doi.org/10.1200/JCO.2018.78.2276
Reck M, Rodríguez-Abreu D, Robinson AG, Hui R, Csőszi T, Fülöp A, Gottfried M, Peled N, Tafreshi A, Cuffe S, O'Brien M, Rao S, Hotta K, Leiby MA, Lubiniecki GM, Shentu Y, Rangwala R, Brahmer JR (2016) KEYNOTE-024 Investigators. Pembrolizumab versus Chemotherapy for PD-L1-Positive Non-Small-Cell Lung Cancer. N Engl J Med 375(19):1823–1833. https://doi.org/10.1056/NEJMoa1606774
Mok TSK, Wu YL, Kudaba I, Kowalski DM, Cho BC, Turna HZ, Castro G Jr, Srimuninnimit V, Laktionov KK, Bondarenko I, Kubota K, Lubiniecki GM, Zhang J, Kush D, Lopes G (2019) KEYNOTE-042 Investigators. Pembrolizumab versus chemotherapy for previously untreated, PD-L1-expressing, locally advanced or metastatic non-small-cell lung cancer (KEYNOTE-042): a randomised, open-label, controlled, phase 3 trial. Lancet. 393(10183):1819–1830. https://doi.org/10.1016/S0140-6736(18)32409-7
Assoun S, Theou-Anton N, Nguenang M, Cazes A, Danel C, Abbar B, Pluvy J, Gounant V, Khalil A, Namour C, Brosseau S, Zalcman G (2019) Association of TP53 mutations with response and longer survival under immune checkpoint inhibitors in advanced non-small-cell lung cancer. Lung Cancer 132:65–71. https://doi.org/10.1016/j.lungcan.2019.04.005
Frost N, Kollmeier J, Vollbrecht C, Grah C, Matthes B, Pultermann D, von Laffert M, Lüders H, Olive E, Raspe M, Mairinger T, Ochsenreither S, Blum T, Hummel M, Suttorp N, Witzenrath M, Grohé C (2021) KRASG12C/TP53 co-mutations identify long-term responders to first line palliative treatment with pembrolizumab monotherapy in PD-L1 high (≥50%) lung adenocarcinoma. Transl Lung Cancer Res. 10(2):737–752. https://doi.org/10.21037/tlcr-20-958.PMID:33718018;PMCID:PMC7947421
Lyu Q, Lin A, Cao M, Xu A, Luo P, Zhang J (2020) Alterations in TP53 Are a Potential Biomarker of Bladder Cancer Patients Who Benefit From Immune Checkpoint Inhibition. Cancer Control 27(1):1073274820976665. https://doi.org/10.1177/1073274820976665
Sun H, Liu SY, Zhou JY, Xu JT, Zhang HK, Yan HH, Huan JJ, Dai PP, Xu CR, Su J, Guan YF, Yi X, Yu RS, Zhong WZ, Wu YL (2020) Specific TP53 subtype as biomarker for immune checkpoint inhibitors in lung adenocarcinoma. EBioMedicine 60:102990. https://doi.org/10.1016/j.ebiom.2020.102990
ClinicalTrials.gov [Internet]. Bethesda (MD): National Library of Medicine (US). 2000 Feb 29 - . Identifier NCT02628067, Study of Pembrolizumab (MK-3475) in Participants With Advanced Solid Tumors (MK-3475-158/KEYNOTE-158); 2015 Dec 9 [cited 2021 Dec 1]; [about 4 screens]. Available from: https://clinicaltrials.gov/ct2/show/record/NCT02628067
Marabelle A, Le DT, Ascierto PA, Di Giacomo AM, De Jesus-Acosta A, Delord JP, Geva R, Gottfried M, Penel N, Hansen AR, Piha-Paul SA, Doi T, Gao B, Chung HC, Lopez-Martin J, Bang YJ, Frommer RS, Shah M, Ghori R, Joe AK, Pruitt SK, Diaz LA Jr (2020) Efficacy of Pembrolizumab in Patients With Noncolorectal High Microsatellite Instability/Mismatch Repair-Deficient Cancer: Results From the Phase II KEYNOTE-158 Study. J Clin Oncol 38(1):1–10. https://doi.org/10.1200/JCO.19.02105
Marabelle A, Fakih M, Lopez J, Shah M, Shapira-Frommer R, Nakagawa K, Chung HC, Kindler HL, Lopez-Martin JA, Miller WH Jr, Italiano A, Kao S, Piha-Paul SA, Delord JP, McWilliams RR, Fabrizio DA, Aurora-Garg D, Xu L, Jin F, Norwood K, Bang YJ (2020) Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab: prospective biomarker analysis of the multicohort, open-label, phase 2 KEYNOTE-158 study. Lancet Oncol 21(10):1353–1365. https://doi.org/10.1016/S1470-2045(20)30445-9
McGrail DJ, Pilié PG, Rashid NU, Voorwerk L, Slagter M, Kok M, Jonasch E, Khasraw M, Heimberger AB, Lim B, Ueno NT, Litton JK, Ferrarotto R, Chang JT, Moulder SL, Lin SY (2021) High tumor mutation burden fails to predict immune checkpoint blockade response across all cancer types. Ann Oncol 32(5):661–672. https://doi.org/10.1016/j.annonc.2021.02.006
Acknowledgements
Not applicable.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
Gabriel Roman Souza and Daruka Mahadevan contributed to the study conception and design. Material preparation and analysis were performed by Gabriel Roman Souza. Data collection was performed by all authors. The first draft of the manuscript was written by Gabriel Roman Souza and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
The institutional review board of the University of Texas Health San Antonio approved this study.
Informed consent
For this type of study, formal consent is not required.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Research involving human participants and/or animals
This research involves human participants. This study was performed in line with the principles of the Declaration of Helsinki. The institutional review board of the University of Texas Health San Antonio approved this study.
Conflict of interests
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Roman Souza, G., Abdalla, A., Arora, S. et al. Real-world data of off-label drug use in patients with actionable genomic alterations on next-generation sequencing. Invest New Drugs 40, 643–649 (2022). https://doi.org/10.1007/s10637-022-01213-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10637-022-01213-x