Abstract
Background/Aims
Aberrant Peroxisomal Biogenesis Factor 26 (PEX26) occurs in multiple cell process. However, the role of PEX26 in colorectal cancer (CRC) development remains unknown. We aimed to study PEX26 expression, regulation, and function in CRC cells.
Methods
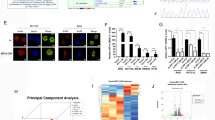
Using the bioinformatic analysis, real-time quantitative PCR, and immunohistochemistry staining, we detected the expression of PEX26 in CRC and normal tissues. We performed functional experiments in vitro to elucidate the effect of PEX26 on CRC cells. We analyzed the RNA-seq data to reveal the downstream regulating network of PEX26.
Results
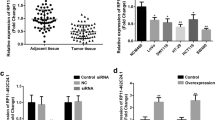
PEX26 is significantly down-regulated in CRC and its low expression correlates with the poor overall survival of CRC patients. We further demonstrated that PEX26 over-expression inhibits the ability of CRC cell migration, invasion, and epithelial–mesenchymal transition (EMT), while PEX26 knockdown promotes the malignant phenotypes of migration, invasion, and EMT via activating the Wnt pathway.
Conclusion
Overall, our results showed that the loss of PEX26 contributes to the malignant phenotype of CRC. PEX26 may serve as a novel metastasis repressor for CRC.






Similar content being viewed by others
References
Xie H, Gong Y, Kuang J et al. Computed-tomography-determined sarcopenia is a useful imaging biomarker for predicting postoperative outcomes in elderly colorectal cancer patients. Cancer Res Treat: Off J Korean Cancer Assoc. 2020;52:957.
Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. 2018;68:394–424.
Tanaka AJ, Okumoto K, Tamura S et al. A newly identified mutation in the PEX26 gene is associated with a milder form of Zellweger spectrum disorder. Cold Spring Harbor Mol Case Stud. 2019. https://doi.org/10.1101/mcs.a003483.
Okugawa Y, Grady WM, Goel A. Epigenetic alterations in colorectal cancer: emerging biomarkers. Gastroenterology 2015;149:1204-1225.e1212.
He H, Chen E, Lei L et al. Alteration of the tumor suppressor SARDH in sporadic colorectal cancer: a functional and transcriptome profiling-based study. Mo Carcinog. 2019;58:957–966.
Chen E, Yang F, He H et al. Alteration of tumor suppressor BMP5 in sporadic colorectal cancer: a genomic and transcriptomic profiling based study. Mol Cancer. 2018;17:176.
Chen L, Pan X, Hu X et al. Gene expression differences among different MSI statuses in colorectal cancer. Int J Cancer. 2018;143:1731–1740.
Wanders RJ. Peroxisomes in human health and disease: metabolic pathways, metabolite transport, interplay with other organelles and signal transduction. Sub-Cell Biochem. 2013;69:23–44.
Wanders RJ. Metabolic functions of peroxisomes in health and disease. Biochimie 2014;98:36–44.
Lodhi IJ, Semenkovich CF. Peroxisomes: a nexus for lipid metabolism and cellular signaling. Cell Metab. 2014;19:380–392.
Fujiki Y. Peroxisome biogenesis and human peroxisome-deficiency disorders. Proc Jpn Acad Ser B, Phys Biol Sci. 2016;92:463–477.
Argyriou C, D’Agostino MD, Braverman N. Peroxisome biogenesis disorders. Transl Sci Rare Dis. 2016;1:111–144.
Santos MJ, Quintanilla RA, Toro A et al. Peroxisomal proliferation protects from beta-amyloid neurodegeneration. J Biol Chem. 2005;280:41057–41068.
Fransen M, Nordgren M, Wang B, Apanasets O. Role of peroxisomes in ROS/RNS-metabolism: implications for human disease. Biochim Biophys Acta. 2012;1822:1363–1373.
Singh I, Singh AK, Contreras MA. Peroxisomal dysfunction in inflammatory childhood white matter disorders: an unexpected contributor to neuropathology. J Child Neurol. 2009;24:1147–1157.
Zhang J, Tripathi DN, Jing J et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat Cell Biol. 2015;17:1259–1269.
Mo S, Dai W, Xiang W et al. Prognostic and predictive value of an autophagy-related signature for early relapse in stages I–III colon cancer. Carcinogenesis 2019;40:861–870.
D’Arcangelo D, Giampietri C, Muscio M, Scatozza F, Facchiano F, Facchiano A. WIPI1, BAG1, and PEX3 autophagy-related genes are relevant melanoma markers. Oxidat Med Cell Longev. 2018;2018:1471682.
Cai M, Sun X, Wang W et al. Disruption of peroxisome function leads to metabolic stress, mTOR inhibition, and lethality in liver cancer cells. Cancer Lett. 2018;421:82–93.
Waterham HR, Ebberink MS. Genetics and molecular basis of human peroxisome biogenesis disorders. Biochim Biophys Acta. 2012;1822:1430–1441.
Chen E, Li Q, Wang H, Yang F, Min L, Yang J. MiR-92a promotes tumorigenesis of colorectal cancer, a transcriptomic and functional based study. Biomed Pharmacother Biomed Pharmacother. 2018;106:1370–1377.
Neuhaus C, Eisenberger T, Decker C et al. Next-generation sequencing reveals the mutational landscape of clinically diagnosed Usher syndrome: copy number variations, phenocopies, a predominant target for translational read-through, and PEX26 mutated in Heimler syndrome. Mol Genet Genomic Med. 2017;5:531–552.
Gao FJ, Hu FY, Xu P et al. Expanding the clinical and genetic spectrum of Heimler syndrome. Orphanet J Rare Dis. 2019;14:290.
Dahabieh MS, Ha Z, Di Pietro E et al. Peroxisomes protect lymphoma cells from HDAC inhibitor-mediated apoptosis. Cell Death Differ. 2017;24:1912–1924.
Wen J, Xiong K, Aili A et al. PEX5, a novel target of microRNA-31-5p, increases radioresistance in hepatocellular carcinoma by activating Wnt/β-catenin signaling and homologous recombination. Theranostics 2020;10:5322–5340.
Nieto MA, Huang RY, Jackson RA, Thiery JP. EMT: 2016. Cell 2016;166:21–45.
Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell 2009;139:871–890.
Dongre A, Weinberg RA. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat Rev Mol Cell Biol. 2019;20:69–84.
Ye X, Tam WL, Shibue T et al. Distinct EMT programs control normal mammary stem cells and tumour-initiating cells. Nature 2015;525:256–260.
Rhim AD, Mirek ET, Aiello NM et al. EMT and dissemination precede pancreatic tumor formation. Cell 2012;148:349–361.
Migita T, Ueda A, Ohishi T et al. Epithelial-mesenchymal transition promotes SOX2 and NANOG expression in bladder cancer. Lab Invest: J Tech Methods Pathol. 2017;97:567.
Yan X, Yan L, Liu S, Shan Z, Tian Y, Jin Z. N-cadherin, a novel prognostic biomarker, drives malignant progression of colorectal cancer. Mol Med Rep. 2015;12:2999–3006.
Aiello NM, Brabletz T, Kang Y, Nieto MA, Weinberg RA, Stanger BZ. Upholding a role for EMT in pancreatic cancer metastasis. Nature 2017;547:E7-e8.
Huang RY, Guilford P, Thiery JP. Early events in cell adhesion and polarity during epithelial-mesenchymal transition. J Cell Sci. 2012;125:4417–4422.
Yilmaz M, Christofori G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009;28:15–33.
Yeung KT, Yang J. Epithelial-mesenchymal transition in tumor metastasis. Mol Oncol. 2017;11:28–39.
Lamouille S, Xu J, Derynck R. Molecular mechanisms of epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2014;15:178–196.
Santiago L, Daniels G, Wang DW, Deng FM, Lee P. Wnt signaling pathway protein LEF1 in cancer, as a biomarker for prognosis and a target for treatment. Am J Cancer Res. 2017;7:1389–1406.
Yang SC, Liu Y, Li MY et al. FOXP3 promotes tumor growth and metastasis by activating Wnt/beta-catenin signaling pathway and EMT in non-small cell lung cancer. Mol Cancer. 2017;16:1–12.
Bugter JM, Fenderico N, Maurice MM. Mutations and mechanisms of WNT pathway tumour suppressors in cancer. Nat Rev Cancer. 2020;21:5–21.
Garcia de Herreros A, Dunach M. Intracellular signals activated by canonical Wnt ligands independent of GSK3 inhibition and beta-catenin stabilization. Cells-Basel 2019;8:1148.
Funding
This study was supported by grants from the National Natural Scientific Foundation of China (No. 81974378 and No. 82003115) and Shaanxi Fundamental Science Research Project for Chemistry & Biology (22JHQ084).
Author information
Authors and Affiliations
Contributions
EC and JY designed and drafted the manuscript. BY, LG, SW, MW, YT, and DY performed the experiments. LC helped analyzed the statistics. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors reported no conflicts of interest in this work.
Ethical approval
The study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Northwest University.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Yan, B., Cao, L., Gao, L. et al. PEX26 Functions as a Metastasis Suppressor in Colorectal Cancer. Dig Dis Sci 69, 112–122 (2024). https://doi.org/10.1007/s10620-023-08168-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-023-08168-w