Abstract
Background
Alcohol-associated liver disease accounts for half of cirrhosis-related deaths worldwide. The spectrum of disease varies from simple steatosis to fibrosis, cirrhosis and ultimately hepatocellular carcinoma. Understanding the disease on a molecular level helps us to develop therapeutic targets.
Aim
We performed transcriptomic analysis in liver and ileum from chronic plus binge ethanol-fed mice, and we assessed the role of selected differentially expressed genes and their association with serum bile acids and gut microbiota.
Methods
Wild-type mice were subjected to a chronic Lieber-DeCarli diet model for 8 weeks followed by one binge of ethanol. RNA-seq analysis was performed on liver and ileum samples. Associations between selected differentially regulated genes and serum bile acid profile or fecal bacterial profiling (16S rDNA sequencing) were investigated.
Results
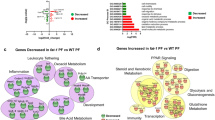
We provide a comprehensive transcriptomic analysis to identify differentially expressed genes, KEGG pathways, and gene ontology functions in liver and ileum from chronic plus binge ethanol-fed mice. In liver, we identified solute carrier organic anion transporter family, member 1a1 (Slco1a1; encoding for organic anion transporting polypeptides (OATP) 1A1), as the most down-regulated mRNA, and it is negatively correlated with serum cholic acid level. Prokineticin 2 (Prok2) mRNA, a cytokine-like molecule associated with gastrointestinal tract inflammation, was significantly down-regulated in ethanol-fed mice. Prok2 mRNA expression was negatively correlated with abundance of Allobaculum (genus), Coprococcus (genus), Lachnospiraceae (family), Lactococcus (genus), and Cobriobacteriaceae (family), while it positively correlated with Bacteroides (genus).
Conclusions
RNA-seq analysis revealed unique transcriptomic signatures in the liver and intestine following chronic plus binge ethanol feeding.






Similar content being viewed by others
Abbreviations
- DEG:
-
Differentially expressed gene
- GO:
-
Gene ontology
- Slco1a1 :
-
Solute carrier organic anion transporter family, member 1a1
- OATP:
-
Organic anion transporting polypeptides
- NTCP:
-
Sodium-dependent taurocholate cotransporting polypeptide
- TNF:
-
Tumor necrosis factor
References
Gao B, Bataller R. Alcoholic liver disease: pathogenesis and new therapeutic targets. Gastroenterology. 2011;141(5):1572–1585. https://doi.org/10.1053/j.gastro.2011.09.002.
Ramond MJ, Poynard T, Rueff B, et al. A randomized trial of prednisolone in patients with severe alcoholic hepatitis. N Engl J Med. 1992;326(8):507–512. https://doi.org/10.1056/NEJM199202203260802.
Saberi B, Dadabhai AS, Jang YY, Gurakar A, Mezey E. Current management of alcoholic hepatitis and future therapies. J Clin Transl Hepatol. 2016;4(2):113–122. https://doi.org/10.14218/JCTH.2016.00006.
Yan AW, Fouts DE, Brandl J, et al. Enteric dysbiosis associated with a mouse model of alcoholic liver disease. Hepatology. 2011;53(1):96–105. https://doi.org/10.1002/hep.24018.
Mutlu EA, Gillevet PM, Rangwala H, et al. Colonic microbiome is altered in alcoholism. Am J Physiol Gastrointest Liver Physiol. 2012;302(9):G966–G978. https://doi.org/10.1152/ajpgi.00380.2011.
Duan Y, Llorente C, Lang S, et al. Bacteriophage targeting of gut bacterium attenuates alcoholic liver disease. Nature. 2019;575(7783):505–511. https://doi.org/10.1038/s41586-019-1742-x.
Brandl K, Hartmann P, Jih LJ, et al. Dysregulation of serum bile acids and FGF19 in alcoholic hepatitis. J Hepatol. 2018;69(2):396–405. https://doi.org/10.1016/j.jhep.2018.03.031.
Chu H, Jiang L, Gao B, et al. The selective PPAR-delta agonist seladelpar reduces ethanol-induced liver disease by restoring gut barrier function and bile acid homeostasis in mice. Transl Res. 2020;. https://doi.org/10.1016/j.trsl.2020.06.006.
Yang AM, Inamine T, Hochrath K, et al. Intestinal fungi contribute to development of alcoholic liver disease. J Clin Investig. 2017;127(7):2829–2841. https://doi.org/10.1172/JCI90562.
Patro R, Duggal G, Love MI, Irizarry RA, Kingsford C. Salmon provides fast and bias-aware quantification of transcript expression. Nat Methods. 2017;14(4):417–419. https://doi.org/10.1038/nmeth.4197.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):550. https://doi.org/10.1186/s13059-014-0550-8.
Yu G, Wang LG, Han Y, He QY. ClusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287. https://doi.org/10.1089/omi.2011.0118.
Hartmann P, Hochrath K, Horvath A, et al. Modulation of the intestinal bile acid/farnesoid X receptor/fibroblast growth factor 15 axis improves alcoholic liver disease in mice. Hepatology. 2018;67(6):2150–2166. https://doi.org/10.1002/hep.29676.
Caporaso JG, Kuczynski J, Stombaugh J, et al. QIIME allows analysis of high-throughput community sequencing data. Nat Methods. 2010;7(5):335–336. https://doi.org/10.1038/nmeth.f.303.
DeSantis TZ, Hugenholtz P, Larsen N, et al. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol. 2006;72(7):5069–5072. https://doi.org/10.1128/AEM.03006-05.
Slijepcevic D, Abbing RLPR, Katafuchi T, et al. Hepatic uptake of conjugated bile acids is mediated by both sodium taurocholate cotransporting polypeptide and organic anion transporting polypeptides and modulated by intestinal sensing of plasma bile acid levels in mice. Hepatology. 2017;66(5):1631–1643. https://doi.org/10.1002/hep.29251.
Martinez-Augustin O, Sanchez de Medina F. Intestinal bile acid physiology and pathophysiology. World J Gastroenterol. 2008;14(37):5630–5640. https://doi.org/10.3748/wjg.14.5630.
Watson RP, Lilley E, Panesar M, et al. Increased prokineticin 2 expression in gut inflammation: role in visceral pain and intestinal ion transport. Neurogastroenterol Motil. 2012;24(1):65–75, e12. https://doi.org/10.1111/j.1365-2982.2011.01804.x.
Pradhan-Sundd T, Vats R, Russell JO, et al. Dysregulated bile transporters and impaired tight junctions during chronic liver injury in mice. Gastroenterology. 2018;155(4):1218–1232.e24. https://doi.org/10.1053/j.gastro.2018.06.048.
Gyongyosi B, Cho Y, Lowe P, et al. Alcohol-induced IL-17A production in Paneth cells amplifies endoplasmic reticulum stress, apoptosis, and inflammasome-IL-18 activation in the proximal small intestine in mice. Mucosal Immunol. 2019;12(4):930–944. https://doi.org/10.1038/s41385-019-0170-4.
Liu H, Gong M, French BA, et al. Aberrant modulation of the BRCA1 and G1/S cell cycle pathways in alcoholic hepatitis patients with Mallory Denk Bodies revealed by RNA sequencing. Oncotarget. 2015;6(40):42491–42503. https://doi.org/10.18632/oncotarget.6382.
Khanova E, Wu R, Wang W, et al. Pyroptosis by caspase11/4-gasdermin-D pathway in alcoholic hepatitis in mice and patients. Hepatology. 2018;67(5):1737–1753. https://doi.org/10.1002/hep.29645.
Liu H, French BA, Li J, Tillman B, French SW. Altered regulation of miR-34a and miR-483-3p in alcoholic hepatitis and DDC fed mice. Exp Mol Pathol. 2015;99(3):552–557. https://doi.org/10.1016/j.yexmp.2015.09.005.
Zhang W, Thompson BJ, Hietakangas V, Cohen SM. MAPK/ERK signaling regulates insulin sensitivity to control glucose metabolism in Drosophila. PLoS Genet. 2011;7(12):e1002429. https://doi.org/10.1371/journal.pgen.1002429.
Kishore R, Hill JR, McMullen MR, Frenkel J, Nagy LE. ERK1/2 and Egr-1 contribute to increased TNF-alpha production in rat Kupffer cells after chronic ethanol feeding. Am J Physiol Gastrointest Liver Physiol. 2002;282(1):G6–G15. https://doi.org/10.1152/ajpgi.00328.2001.
van de Steeg E, Wagenaar E, van der Kruijssen CM, et al. Organic anion transporting polypeptide 1a/1b-knockout mice provide insights into hepatic handling of bilirubin, bile acids, and drugs. J Clin Investig. 2010;120(8):2942–2952. https://doi.org/10.1172/jci42168.
Bull-Otterson L, Feng W, Kirpich I, et al. Metagenomic analyses of alcohol induced pathogenic alterations in the intestinal microbiome and the effect of Lactobacillus rhamnosus GG treatment. PloS One. 2013;8(1):e53028. https://doi.org/10.1371/journal.pone.0053028.
Le Roy T, Llopis M, Lepage P, et al. Intestinal microbiota determines development of non-alcoholic fatty liver disease in mice. Gut. 2013;62(12):1787–1794. https://doi.org/10.1136/gutjnl-2012-303816.
Seitz HK, Bataller R, Cortez-Pinto H, et al. Alcoholic liver disease. Nat Rev Dis Primers. 2018;4(1):16. https://doi.org/10.1038/s41572-018-0014-7.
Acknowledgments
This study was supported in part by NIH Grants R01 AA24726, R01 AA020703, U01 AA026939, by Award Number BX004594 from the Biomedical Laboratory Research & Development Service of the VA Office of Research and Development (to B.S.). The study was also supported by the NIDDK-funded San Diego Digestive Diseases Research Center (P30 DK120515) and by the NIAAA-funded Southern California Research Center for ALPD and Cirrhosis (P50 AA011999).
Author information
Authors and Affiliations
Contributions
LJ was responsible for acquisition, analysis and interpretation of data, and writing of the manuscript; HC performed mouse studies; BG and SL provided assistance in data analysis; YD and YW provided assistance in data acquisition; BS was responsible for the study concept and design, critical revision of the manuscript, and study supervision.
Corresponding author
Ethics declarations
Conflict of interest
B.S. has been consulting for Ferring Research Institute, Intercept Pharmaceuticals, HOST Therabiomics and Patara Pharmaceuticals. B.S.’s institution UC San Diego has received Grant support from BiomX, NGM Biopharmaceuticals, CymaBay Therapeutics, Synlogic Operating Company and Axial Biotherapeutics.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Jiang, L., Chu, H., Gao, B. et al. Transcriptomic Profiling Identifies Novel Hepatic and Intestinal Genes Following Chronic Plus Binge Ethanol Feeding in Mice. Dig Dis Sci 65, 3592–3604 (2020). https://doi.org/10.1007/s10620-020-06461-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-020-06461-6