Abstract
Background
Long non-coding RNAs (lncRNAs) have been increasingly uncovered to participate in multiple human cancers, including pancreatic cancer (PC). However, the underlying mechanisms of most of the lncRNAs have not been fully understood yet.
Aims
In this study, we probed the role and latent mechanism of LINC01420 in PC.
Methods
Several online tools were applied. Gene expression was evaluated by qRT-PCR or Western blot. Both in vitro and in vivo assays were conducted to probe LINC01420 function in PC. ChIP, RIP, and luciferase reporter assays were performed to determine relationships between genes.
Results
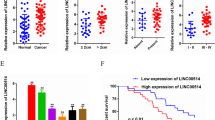
The bioinformatics analyses revealed LINC01420 was highly expressed in PC tissues. Besides, LINC01420 was pronouncedly upregulated in PC cell lines and its depletion controlled PC cell proliferation and EMT in vitro and hindered tumor growth in vivo. Importantly, KRAS was proved to mediate LINC01420-facilitated PC cell proliferation. Further, we explained that KRAS transcription was regulated by MYC, while LINC01420 enhanced the binding of MYC to KRAS promoter in the nucleus of PC cells. Intriguingly, LINC01420 boosted MYC expression in the cytoplasm of PC cells by sponging miR-494-3p.
Conclusion
This study illustrated that LINC01420 accelerates PC progression through releasing miR-494-3p-silenced MYC in cytoplasm and upregulating MYC-activated KRAS in nucleus, unveiling LINC01420 as a latent therapeutic strategy for PC patients.






Similar content being viewed by others
References
Murr MM, Sarr MG, Oishi AJ, van Heerden JA. Pancreatic cancer. CA Cancer J Clin. 1994;44:304–318.
Li D, Xie K, Wolff R, Abbruzzese JL. Pancreatic cancer. Lancet. 2004;363:1049–1057.
Wolfgang CL, Herman JM, Laheru DA, et al. Recent progress in pancreatic cancer. CA Cancer J Clin. 2013;63:318–348.
Yachida S, Jones S, Bozic I, et al. Distant metastasis occurs late during the genetic evolution of pancreatic cancer. Nature. 2010;467:1114–1117.
Jooste V, Dejardin O, Bouvier V, et al. Pancreatic cancer: wait times from presentation to treatment and survival in a population-based study. Int J Cancer. 2016;139:1073–1080.
Cheetham SW, Gruhl F, Mattick JS, Dinger ME. Long noncoding RNAs and the genetics of cancer. Br J Cancer. 2013;108:2419–2425.
Noh JH, Kim KM, McClusky WG, Abdelmohsen K, Gorospe M. Cytoplasmic functions of long noncoding RNAs. Wiley Interdiscip Rev RNA IF4.838. 2018;9:e1471.
Batista PJ, Chang HY. Long noncoding RNAs: cellular address codes in development and disease. Cell. 2013;152:1298–1307.
Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64.
Noh JH, Gorospe M. AKTions by cytoplasmic lncRNA CASC9 promote hepatocellular carcinoma survival. Hepatology (Baltimore, Md.). 2018;68:1675–1677.
Zhang E, Han L, Yin D, et al. H3K27 acetylation activated-long non-coding RNA CCAT1 affects cell proliferation and migration by regulating SPRY4 and HOXB13 expression in esophageal squamous cell carcinoma. Nucleic Acids Res. 2017;45:3086–3101.
Lin A, Li C, Xing Z, et al. The LINK-A lncRNA activates normoxic HIF1α signalling in triple-negative breast cancer. Nat Cell Biol. 2016;18:213–224.
Li GY, Wang W, Sun JY, et al. Long non-coding RNAs AC026904.1 and UCA1: a “one-two punch” for TGF-β-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics. 2018;8:2846–2861.
Liang Y, Chen X, Wu Y, et al. LncRNA CASC9 promotes esophageal squamous cell carcinoma metastasis through upregulating LAMC2 expression by interacting with the CREB-binding protein. Cell Death Differ. 2018;25:1980–1995.
Yang L, Tang Y, He Y, et al. High expression of LINC01420 indicates an unfavorable prognosis and modulates cell migration and invasion in nasopharyngeal carcinoma. J Cancer. 2017;8:97–103.
Mallakin A, Taneja P, Matise LA, Willingham MC, Inoue K. Expression of Dmp1 in specific differentiated, nonproliferating cells and its regulation by E2Fs. Oncogene. 2006;25:7703–7713.
Lo HC, Zhang XHF. EMT in metastasis: finding the right balance. Dev Cell. 2018;45:663–665.
Mongroo PS, Noubissi FK, Cuatrecasas M, et al. IMP-1 displays cross-talk with K-Ras and modulates colon cancer cell survival through the novel proapoptotic protein CYFIP2. Cancer Res. 2011;71:2172–2182.
Wang Z, Yang B, Zhang M, et al. lncRNA epigenetic landscape analysis identifies EPIC1 as an oncogenic lncRNA that interacts with MYC and promotes cell-cycle progression in cancer. Cancer Cell. 2018;33:706–720.
Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
Knoll M, Lodish HF, Sun L. Long non-coding RNAs as regulators of the endocrine system. Nat Rev Endocrinol. 2015;11:151–160.
Deng S-J, Chen H-Y, Zeng Z, et al. Nutrient stress-dysregulated antisense lncRNA GLS-AS impairs GLS-mediated metabolism and represses pancreatic cancer progression. Cancer Res. 2019;79:1398–1412.
Deng S-J, Chen H-Y, Ye Z, et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene. 2018;37:5811–5828.
Fu Z, Chen C, Zhou Q, et al. LncRNA HOTTIP modulates cancer stem cell properties in human pancreatic cancer by regulating HOXA9. Cancer Lett. 2017;410:68–81.
Slebos RJ, Kibbelaar RE, Dalesio O, et al. K-ras oncogene activation as a prognostic marker in adenocarcinoma of the lung. N Engl J Med. 1990;323:561–565.
Ichikawa Y, Nishida M, Suzuki H, et al. Mutation of K-ras protooncogene is associated with histological subtypes in human mucinous ovarian tumors. Cancer Res. 1994;54:33–35.
Abubaker J, Bavi P, Al-Haqawi W, et al. Prognostic significance of alterations in KRAS isoforms KRAS-4A/4B and KRAS mutations in colorectal carcinoma. J Pathol. 2009;219:435–445.
Ying H, Kimmelman AC, Lyssiotis CA, et al. Oncogenic Kras maintains pancreatic tumors through regulation of anabolic glucose metabolism. Cell. 2012;149:656–670.
Iacobuzio-Donahue CA, Herman JM. Autophagy, p53, and pancreatic cancer. N Engl J Med. 2014;370:1352–1353.
Mueller S, Engleitner T, Maresch R, et al. Evolutionary routes and KRAS dosage define pancreatic cancer phenotypes. Nature. 2018;554:62–68.
Kamerkar S, LeBleu VS, Sugimoto H, et al. Exosomes facilitate therapeutic targeting of oncogenic KRAS in pancreatic cancer. Nature. 2017;546:498–503.
Zhang L, Yu S, Wang C, Jia C, Lu Z, Chen J. Establishment of a non-coding RNAomics screening platform for the regulation of KRAS in pancreatic cancer by RNA sequencing. Int J Oncol. 2018;53:2659–2670.
Li X, Deng S-J, Zhu S, et al. Hypoxia-induced lncRNA-NUTF2P3-001 contributes to tumorigenesis of pancreatic cancer by derepressing the miR-3923/KRAS pathway. Oncotarget. 2016;7:6000–6014.
Liu P, Yang H, Zhang J, et al. The lncRNA MALAT1 acts as a competing endogenous RNA to regulate KRAS expression by sponging miR-217 in pancreatic ductal adenocarcinoma. Sci Rep. 2017;7:5186.
Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505:344–352.
Kim WK, Park M, Kim Y-K, et al. MicroRNA-494 downregulates KIT and inhibits gastrointestinal stromal tumor cell proliferation. Clin Cancer Res. 2011;17:7584–7594.
Zhan M-N, Yu X-T, Tang J, et al. MicroRNA-494 inhibits breast cancer progression by directly targeting PAK1. Cell Death Dis. 2017;8:e2529–e2529.
Lim L, Balakrishnan A, Huskey N, et al. MicroRNA-494 within an oncogenic microRNA megacluster regulates G1/S transition in liver tumorigenesis through suppression of mutated in colorectal cancer. Hepatology (Baltimore, Md.). 2014;59:202–215.
Li L, Li Z, Kong X, et al. Down-regulation of MicroRNA-494 via loss of SMAD4 increases FOXM1 and β;-catenin signaling in pancreatic ductal adenocarcinoma cells. Gastroenterology. 2014;147:485–497.
Acknowledgment
We appreciate all the contributors during our study.
Funding
None.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1 Mechanistic investigation in SW1990 and HPAC cells.
(A) The binding of MYC protein to KRAS promoter in SW1990 and HPAC cells was validated by ChIP assay. (B) The interaction between LINC01420 and MYC protein in these two PC cells was proved by RIP assay. (C-D) ChIP assay uncovered the inhibition of LINC01420 silence on MYC occupation in KRAS promoter, (E-F) qRT-PCR result of MYC expression in SW1990 and HPAC cells under diverse transfections. * P < 0.05, ** P < 0.01. (TIFF 8015 kb)
Rights and permissions
About this article
Cite this article
Zhai, H., Zhang, X., Sun, X. et al. Long Non-coding RNA LINC01420 Contributes to Pancreatic Cancer Progression Through Targeting KRAS Proto-oncogene. Dig Dis Sci 65, 1042–1052 (2020). https://doi.org/10.1007/s10620-019-05829-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-019-05829-7