Abstract
Background
DNA mismatch repair (MMR) deficiency results in a strong mutator phenotype and high-frequency microsatellite instability (MSI-H), which are the hallmarks of many tumors.
Aim
The objective of this study is to investigate the promoter CpG island methylation status of mismatch repair genes human mutL homolog 1 (hMLH1), human mutS homolog 2 (hMSH2), and O6-methylguanine-DNA methyltransferase (MGMT) in esophageal squamous cell carcinoma (ESCC) and its roles in alkylating agents chemotherapy.
Methods
Real-time methylation-specific polymerase chain reaction (PCR) (real-time MSP) was employed to detect promoter CpG island methylation of the hMLH1, hMSH2, as well as MGMT genes in 235 surgical tumor tissue samples from ESCC patients and their corresponding normal tissue samples.
Results
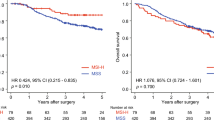
Promoter CpG island methylation of hMLH1, hMSH2, and MGMT were detectable in 43.4, 28.9, and 40.4% of ESCC tumor DNA, respectively, and the loss rates of hMLH1, hMSH2, and MGMT protein expression were 48.6, 34.5, and 40.9% in tumor tissues, respectively. For the entire population of 235 ESCC patients who were enrolled in operating treatment combined with radiotherapy and chemotherapy with alkylating agents, there was a significant difference in the overall survival between patients with methylated MGMT promoter and those with an unmethylated MGMT promoter (P < 0.05).
Conclusion
Promoter CpG island methylation may be a frequent event in ESCC carcinogenesis. Detection of the methylated sequences of hMLH1, hMSH2, and MGMT appears to be promising as a predictive factor in primary ESCC.




Similar content being viewed by others
References
Naidoo R, Ramburan A, Reddi A, Chetty R. Aberrations in the mismatch repair genes and the clinical impact on oesophageal squamous carcinomas from a high incidence area in South Africa. J Clin Pathol. 2005;58:281–284.
Enzinger PC, Mayer RJ. Esophageal cancer. N Engl J Med. 2003;349:2241–2252.
Qiao YL, Wang GQ, Dawsey SM. Esophageal cancer in North Central China. World Gastroenterol News. 2005;10:27–28.
Guohong Z, Min S, Duenmei W, Songnian H, Min L, Jinsong L, Hongbin L, Feng Z, Dongping T, Heling Y, Zhicai L, Shiyong L, Quansheng G, Xiaoyun L, Yuxia G. Genetic heterogeneity of oesophageal cancer in high-incidence areas of southern and northern China. PLoS One. 2010;5:e9668.
An JY, Fan ZM, Gao SS, Zhuang ZH, Qin YR, Li JL, He X, Tsao GS, Wang LD. Loss of heterozygosity in multistage carcinogenesis of esophageal carcinoma at high-incidence area in Henan Province, China. World J Gastroenterol. 2005;11:2055–2060.
Guo XQ, Wang SJ, Zhang LW, Wang XL, Zhang JH, Guo W. DNA methylation and loss of protein expression in esophageal squamous cell carcinogenesis of high-risk area. J Exp Clin Cancer Res. 2007;26:587–594.
Li GM. Mechanisms and functions of DNA mismatch repair. Cell Res. 2008;18:85–98.
Fukui K. DNA mismatch repair in eukaryotes and bacteria. J Nucleic Acids. 2010;2010. pii: 260512.
Kawaguchi M, Banno K, Yanokura M, Kobayashi Y, Kishimi A, Ogawa S, Kisu I, Nomura H, Hirasawa A, Susumu N, Aoki D. Analysis of candidate target genes for mononucleotide repeat mutation in microsatellite instability-high (MSI-H) endometrial cancer. Int J Oncol. 2009;35:977–982.
D’Errico M, de Rinaldis E, Blasi MF, Viti V, Falchetti M, Calcagnile A, Sera F, Saieva C, Ottini L, Palli D, Palombo F, Giuliani A, Dogliotti E. Genome-wide expression profile of sporadic gastric cancers with microsatellite instability. Eur J Cancer. 2009;45:461–469.
Vageli D, Daniil Z, Dahabreh J, Karagianni E, Vamvakopoulou DN, Ioannou MG, Scarpinato K, Vamvakopoulos NC, Gourgoulianis KI, Koukoulis GK. Phenotypic mismatch repair hMSH2 and hMLH1 gene expression profiles in primary non-small cell lung carcinomas. Lung Cancer. 2009;64:282–288.
Conde J, Silva SN, Azevedo AP, Teixeira V, Pina JE, Rueff J, Gaspar JF. Association of common variants in mismatch repair genes and breast cancer susceptibility: A multigene study. BMC Cancer. 2009;9:344.
Stasikowska-Kanicka O, Stawerski P, Wagrowska-Danilewicz M, Danilewicz M. Immunohistochemical analysis of hMLH1 and hMSH2 proteins in serous ovarian tumours. Pol J Pathol. 2009;60:174–178.
Zekri AR, Sabry GM, Bahnassy AA, Shalaby KA, Abdel-Wahabh SA, Zakaria S. Mismatch repair genes (hMLH1, hPMS1, hPMS2, GTBP/hMSH6, hMSH2) in the pathogenesis of hepatocellular carcinoma. World J Gastroenterol. 2005;11:3020–3026.
Shah SN, Hile SE, Eckert KA. Defective mismatch repair, microsatellite mutation bias, and variability in clinical cancer phenotypes. Cancer Res. 2010;70:431–435.
Yan HL, Hao LQ, Jin HY, Xing QH, Xue G, Mei Q, He J, He L, Sun SH. Clinical features and mismatch repair genes analyses of Chinese suspected hereditary non-polyposis colorectal cancer: a cost-effective screening strategy proposal. Cancer Sci. 2008;99:770–780.
Seifert M, Reichrath J. The role of the human DNA mismatch repair gene hMSH2 in DNA repair, cell cycle control and apoptosis: implications for pathogenesis, progression and therapy of cancer. J Mol Histol. 2006;37:301–307.
Lynch PM. The hMSH2 and hMLH1 genes in hereditary nonpolyposis colorectal cancer. Surg Oncol Clin N Am. 2009;18:611–624.
Arai T, Kasahara I, Sawabe M, Honma N, Aida J, Tabubo K. Role of methylation of the hMLH1 gene promoter in the development of gastric and colorectal carcinoma in the elderly. Geriatr Gerontol Int. 2010;10:S207–S212.
Kim HG, Lee S, Kim DY, Ryu SY, Joo JK, Kim JC, Lee KH, Lee JH. Aberrant methylation of DNA mismatch repair genes in elderly patients with sporadic gastric carcinoma: A comparison with younger patients. J Surg Oncol. 2010;101:28–35.
Czerninski R, Krichevsky S, Ashhab Y, Gazit D, Patel V, Ben-Yehuda D. Promoter hypermethylation of mismatch repair genes, hMLH1 and hMSH2 in oral squamous cell carcinoma. Oral Dis. 2009;15:206–213.
Gu M, Kim D, Bae Y, Choi J, Kim S, Song S. Analysis of microsatellite instability, protein expression and methylation status of hMLH1 and hMSH2 genes in gastric carcinomas. Hepatogastroenterology. 2009;56:899–904.
Kang SH, Park KJ, Kim CY, Yu MO, Park CK, Park SH, Chung YG. O(6)-methylguanine DNA methyltransferase status determined by promoter methylation and immunohistochemistry in gliosarcoma and their clinical implications. J Neurooncol. 2010;101:477–486.
Metellus P, Coulibaly B, Nanni I, Fina F, Eudes N, Giorgi R, Barrie M, Chinot O, Fuentes S, Dufour H, Ouafik L, Figarella-Branger D. Prognostic impact of O6-methylguanine-DNA methyltransferase silencing in patients with recurrent glioblastoma multiforme who undergo surgery and carmustine wafer implantation: a prospective patient cohort. Cancer. 2009;115:4783–4794.
International Union Against Cancer (UICC). In: Sobin LH, Gospodarowicz MK, Wittekind Ch, eds. TNM Classification of Malignant Tumours, 7th ed. New York: Wiley-Liss; 2010.
Nie Y, Liao J, Zhao X, Song Y, Yang GY, Wang LD, Yang CS. Detection of multiple gene hypermethylation in the development of esophageal squamous cell carcinoma. Carcinogenesis. 2002;23:1713–1720.
Kawaguchi K, Oda Y, Saito T, Yamamoto H, Takahira T, Kobayashi C, Tamiya S, Tateishi N, Iwamoto Y, Tsuneyoshi M. DNA hypermethylation status of multiple genes in soft tissue sarcomas. Mod Pathol. 2006;19:106–114.
Ling ZQ, Tanaka A, Li P, Nakayama T, Fujiyama Y, Hattori T, Sugihara H. Microsatellite instability with promoter methylation and silencing of hMLH1 can regionally occur during progression of gastric carcinoma. Cancer Lett. 2010;297:244–251.
Cai Y, Wu MH, Ludeman SM, Grdina DJ, Dolan ME. Role of O6-alkylguanine-DNA alkyltransferase in protecting against cyclophosphamide-induced toxicity and mutagenicity. Cancer Res. 1999;59:3059–3063.
van Nifterik KA, van den Berg J, van der Meide WF, Ameziane N, Wedekind LE, Steenbergen RD, Leenstra S, Lafleur MV, Slotman BJ, Stalpers LJ, Sminia P. Absence of the MGMT protein as well as methylation of the MGMT promoter predict the sensitivity for temozolomide. Br J Cancer. 2010;103:29–35.
Fukushima T, Takeshima H, Kataoka H. Anti-glioma therapy with temozolomide and status of the DNA-repair gene MGMT. Anticancer Res. 2009;29:4845–4854.
Martin L, Marples B, Coffey M, Lawler M, Hollywood D, Marignol L. Recognition of O6MeG lesions by MGMT and mismatch repair proficiency may be a prerequisite for low-dose radiation hypersensitivity. Radiat Res. 2009;172:405–413.
Sardi I, Cetica V, Massimino M, Buccoliero AM, Giunti L, Genitori L, Aricò M. Promoter methylation and expression analysis of MGMT in advanced pediatric brain tumors. Oncol Rep. 2009;22:773–779.
Zhao H, Pestina TI, Nasimuzzaman M, Mehta P, Hargrove PW, Persons DA. Amelioration of murine beta-thalassemia through drug selection of hematopoietic stem cells transduced with a lentiviral vector encoding both gamma-globin and the MGMT drug-resistance gene. Blood. 2009;113:5747–5756.
Kitange GJ, Carlson BL, Mladek AC, Decker PA, Schroeder MA, Wu W, Grogan PT, Giannini C, Ballman KV, Buckner JC, James CD, Sarkaria JN. Evaluation of MGMT promoter methylation status and correlation with temozolomide response in orthotopic glioblastoma xenograft model. J Neurooncol. 2009;92:23–31.
Rivera AL, Pelloski CE, Gilbert MR, Colman H, De La Cruz C, Sulman EP, Bekele BN, Aldape KD. MGMT promoter methylation is predictive of response to radiotherapy and prognostic in the absence of adjuvant alkylating chemotherapy for glioblastoma. Neuro Oncol. 2010;12:116–121.
Acharya S, Foster PL, Brooks P, Fishel R. The coordinated functions of the E. coli MutS and MutL proteins in mismatch repair. Mol Cell. 2003;12:233–246.
Schmutte C, Sadoff MM, Shim KS, Acharya S, Fishel R. The interaction of DNA mismatch repair proteins with human exonuclease I. J Biol Chem. 2001;276:33011–33018.
Burgart LJ. Testing for defective DNA mismatch repair in colorectal carcinoma: a practical guide. Arch Pathol Lab Med. 2005;129:1385–1389.
Yan T, Schupp JE, Hwang HS, Wagner MW, Berry SE, Strickfaden S, Veigl ML, Sedwick WD, Boothman DA, Kinsella TJ. Loss of DNA mismatch repair imparts defective cdc2 signaling and G(2) arrest responses without altering survival after ionizing radiation. Cancer Res. 2001;61:8290–8297.
O’Brien V, Brown R. Signalling cell cycle arrest and cell death through the MMR System. Carcinogenesis. 2006;27:682–692.
Feitsma H, Akay A, Cuppen E. Alkylation damage causes MMR-dependent chromosomal instability in vertebrate embryos. Nucleic Acids Res. 2008;36:4047–4056.
Sarkaria JN, Kitange GJ, James CD, Plummer R, Calvert H, Weller M, Wick W. Mechanisms of chemoresistance to alkylating agents in malignant glioma. Clin Cancer Res. 2008;14:2900–2908.
Liu R, Yin LH, Pu YP. Reduced expression of human DNA repair genes in esophageal squamous-cell carcinoma in China. J Toxicol Environ Health A. 2007;70:956–963.
Chang JW, Chen YC, Chen CY, Chen JT, Chen SK, Wang YC. Correlation of genetic instability with mismatch repair protein expression and p53 mutations in non-small cell lung cancer. Clin Cancer Res. 2000;6:1639–1646.
Hayashi M, Tamura G, Jin Z, Kato I, Sato M, Shibuya Y, Yang S, Motoyama T. Microsatellite instability in esophageal squamous cell carcinoma is not associated with hMLH1 promoter hypermethylation. Pathol Int. 2003;53:270–276.
Conde-Pérezprina JC, Luna-López A, López-Diazguerrero NE, Damián-Matsumura P, Zentella A, Königsberg M. Msh2 promoter region hypermethylation as a marker of aging-related deterioration in old retired female breeder mice. Biogerontology. 2008;9:325–334.
Casorelli I, Russo MT, Bignami M. Role of mismatch repair and MGMT in response to anticancer therapies. Anticancer Agents Med Chem. 2008;8:368–380.
Pors K, Patterson LH. DNA mismatch repair deficiency, resistance to cancer chemotherapy and the development of hypersensitive agents. Curr Top Med Chem. 2005;5:1133–1149.
Acknowledgments
This research was partly supported by two grants from the Natural Science Foundation of Zhejiang Province, China (No. Y2080749 and No. Y2091110), a grant from the Zhejiang Province Science and Technology Fund for excellent returnee (No. 2008004), a grant from the Science and Technology General Project of Zhejiang Province (No. 2009C33143), a grant from the Ministry of Education Science and Technology Fund for Excellent Returnee, China (No. 2010609), a grant from the Scientific and Technological Innovations Fund of Henan Province Higher Education (No. 2009HAST1T001), and a grant from the Science and Technology Key Project of the Ministry of Education, China (No. 210130).
Conflicts of interest
The authors declare that they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ling, ZQ., Li, P., Ge, MH. et al. Aberrant Methylation of Different DNA Repair Genes Demonstrates Distinct Prognostic Value for Esophageal Cancer. Dig Dis Sci 56, 2992–3004 (2011). https://doi.org/10.1007/s10620-011-1774-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10620-011-1774-z