Abstract
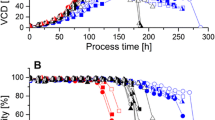
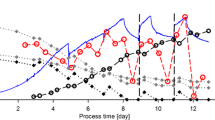
By means of a model predictive control strategy it was possible to ensure a high batch-to-batch reproducibility in animal cell (CHO-cell) suspensions cultured for a recombinant therapeutic protein (EPO) production. The general control objective was derived by identifying an optimal specific growth rate taking productivity, protein quality and process controllability into account. This goal was approached indirectly by controlling the oxygen mass consumed by the cells which is related to specific biomass growth rate and cell concentration profile by manipulating the glutamine feed rate. Process knowledge represented by a classical model was incorporated into the model predictive control algorithm. The controller was employed in several cultivation experiments. During these cultivations, the model parameters were adapted after each sampling event to cope with changes in the process’ dynamics. The ability to predict the state variables, particularly for the oxygen consumption, led to only moderate changes in the desired optimal operational trajectories. Hence, nearly identical oxygen consumption profiles, cell and protein titers as well as sialylation patterns were obtained for all cultivation runs.










Similar content being viewed by others
Abbreviations
- IVC:
-
Integral of viable cells (109 cells h)
- XV :
-
Viable cell density (109 cells L−1)
- Xt :
-
Total cell density (109 cells L−1)
- Glc:
-
Glucose concentration (mM)
- Gln:
-
Glutamine concentration (mM)
- Lac:
-
Lactate concentration (mM)
- NH4 :
-
Ammonia concentration (mM)
- P:
-
Protein (mg)
- W:
-
Culture weight (kg)
- F:
-
Overall feed rate (kg h−1)
- FBase :
-
Base consumption rate (kg h−1)
- FEvap :
-
Evaporation rate (kg h−1)
- FGlc :
-
Glucose feed rate (kg h−1)
- FGln :
-
Glutamine feed rate (kg h−1)
- FSample :
-
Sampling rate (kg h−1)
- XV0 :
-
Viable cell density at start of feeding (109 cells L−1)
- W0 :
-
Culture weight at start of feeding (kg)
- Sf :
-
Substrate feed concentration (mM)
- S:
-
Substrate concentration (mM)
- ρ:
-
Liquid density (kg L−1)
- t:
-
Time (h)
- t0 :
-
Start of feeding (h)
- x:
-
Viable cells (109 cells)
- μ:
-
Specific growth rate (h−1)
- μnet :
-
Net specific growth rate μnet = μ − kd (h−1)
- μset :
-
Setpoint specific growth rate (h−1)
- qi :
-
Specific consumption/production rate of state variable i (mmol 109 cells−1 h−1)
- qGlcmax :
-
Maximum specific glucose consumption rate (mmol 109 cells−1 h−1)
- qGlnmax :
-
Maximum specific glutamine consumption rate (mmol 109 cells−1 h−1)
- kd :
-
Cell decay rate (h−1)
- kdmax :
-
Maximum cell decay rate (h−1)
- kdeg :
-
Degradation constant for glutamine kdeg = 0.004 (h−1)
- YSX :
-
Yield consumed Substrate per cells formed (mmol 109 cells−1 h−1)
- YGlcX :
-
Yield glucose consumed per cells formed (mmol 109 cells−1)
- YGlnX :
-
Yield glutamine consumed per cells formed (mmol 109 cells−1 h−1)
- YLacGlc :
-
Yield lactate formed per glucose consumed (mmol mmol−1)
- \( {\text{Y}}_{{{\text{NH}}_{4}{\rm X}}} \) :
-
Yield ammonia formed per cells formed (mmol 109 cells−1 h−1)
- YOGlc :
-
Yield oxygen consumed per glucose consumed (mmol mmol−1)
- YOGln :
-
Yield oxygen consumed per glutamine consumed (mmol mmol−1)
- KGlc :
-
Glucose saturation constant (mM)
- KGln :
-
Glutamine saturation constant (mM)
- KLac :
-
Lactate inhibition constant for lactate (mM)
- \( {\text{K}}_{{{\text{NH}}_{ 4}^{ + } }} \) :
-
Ammonia inhibition constant for ammonia (mM)
- KO :
-
Inhibition constant (mM)
- k1 :
-
Constant 1 for glucose (mM)
- k2 :
-
Constant 2 for glucose (mM)
- mGln :
-
Glutamine maintenance constant (mmol 109 cells−1 h−1)
- mO :
-
Oxygen maintenance constant (mmol 109 cells−1 h−1)
- \( {\text{n}}_{{{\text{O}}_{ 2} }} \) :
-
Amount of oxygen consumed (mmol)
- mAA :
-
Specific ammonia production rate at low glutamine concentrations (mmol 109 cells−1 h−1) (Zeng et al. 1998)
- OUR:
-
Oxygen uptake rate (mmol L−1 h−1)
- tOUR:
-
Total oxygen uptake rate (mmol h−1)
- tcOUR:
-
\( ={\text{n}}_{{{\text{O}}_{ 2} }} \) total cumulative oxygen uptake rate (mmol)
References
Aehle M, Kuprijanov A, Schaepe S, Simutis R, Lübbert A (2011a) Increasing batch-to-batch reproducibility of CHO cultures by robust open-loop control. Cytotechnology 63:41–47
Aehle M, Schaepe S, Kuprijanov A, Simutis R, Lübbert A (2011b) Simple and efficient control of CHO cell cultures. J Biotechnol 153:56–61
Bork K, Reutter W, Weidemann W, Horstkorte R (2007) Enhanced sialylation of EPO by overexpression of UDP-GlcNAc 2-epimerase/ManAc kinase containing a sialuria mutation in CHO cells. FEBS Lett 581:4195–4198
Camacho EF, Bordons C (1995) Model predictive control in the process industry. Springer, London
Frahm B, Lane P, Atzert H, Munack A, Hoffmann M, Hass VC, Pörtner R (2002) Adaptive, model-based control by the open-loop-feedback-optimal (OLFO) controller for the effective fed-batch cultivation of hybridoma cells. Biotechnol Prog 18:1095–1103
Jenzsch M, Simutis R, Lübbert A (2004) Application of model predictive control to cultivation processes for protein production with genetically modified bacteria. In: Proceedings of the 9th international IFAC symposium, Nancy, pp 511–516
Pirt SJ (1975) Principles of microbe and cell cultivation. Blackwell Scientific Publications, Oxford
Siegwart P, Côté J, Male K (1999) Adaptive control at low glucose concentration of HEK-293 cell serum-free cultures. Biotechnol Prog 15:608–616
Wang MD, Yang M, Huzel N, Butler M (2002) Erythropoietin production from CHO cells grown by continuous culture in a fluidized-bed reactor. Biotechnol Bioeng 77:194–203
Yüzgeç U, Palazoglu A, Romagnoli JA (2010) Refinery scheduling of crude oil unloading, storage and processing using a model predictive control strategy. Comput Chem Eng 34:1671–1686
Zeng A-P, Hu W-S, Deckwer W-D (1998) Variation of stoichiometric ratios and their correlation for monitoring and control of animal cell cultures. Biotechnol Prog 14:434–441
Zhang BJ, Hua B (2007) Effective MILP model for oil refinery-wide production planning and better energy utilization. J Clean Prod 15:439–448
Acknowledgments
The financial support by the Ministry of Science and Education by means of the Excellence Initiative Sachsen-Anhalt is gratefully acknowledged.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Aehle, M., Bork, K., Schaepe, S. et al. Increasing batch-to-batch reproducibility of CHO-cell cultures using a model predictive control approach. Cytotechnology 64, 623–634 (2012). https://doi.org/10.1007/s10616-012-9438-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10616-012-9438-1