Abstract
The McCloud River Redband Trout (MRRT; Oncorhynchus mykiss stonei) is a unique subspecies of rainbow trout that inhabits the isolated Upper McCloud River of Northern California. A major threat to MRRT is introgressive hybridization with non-native rainbow trout from historical stocking and contemporary unauthorized introductions. To help address this concern, we collected RAD-sequencing data on 308 total individuals from MRRT and other California O. mykiss populations and examined population structure using Principal Component and admixture analyses. Our results are consistent with previous studies; we found that populations of MRRT in Sheepheaven, Swamp, Edson, and Moosehead creeks are nonintrogressed. Additionally, we saw no evidence of introgression in Dry Creek, and suggest further investigation to determine if it can be considered a core MRRT conservation population. Sheepheaven Creek was previously thought to be the sole historical lineage of MRRT, but our analysis identified three: Sheepheaven, Edson, and Dry creeks, all of which should be preserved. Finally, we discovered diagnostic and polymorphic SNP markers for monitoring introgression and genetic diversity in MRRT. Collectively, our results provide a valuable resource for the conservation and management of MRRT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The field of conservation genetics aims to reduce the risk of extinction in threatened species. Often the main goal in the management of fish populations is to maintain genetic diversity within and between populations (Ryman et al. 1991; Meffe 1986). Populations within a species that are adapted to different environments display genetic differentiation as a result of their differential adaptations. When the different populations interbreed, the resulting hybrid and/or their offspring may be maladapted. Thus, if populations within a species show genetic and ecological differentiation, they need to be considered separately for conservation management (Small et al. 1998; Frankham et al. 2009). Hence, investigating population genetic structure can provide valuable information to assist with developing conservation strategies for a species, subspecies, and populations.
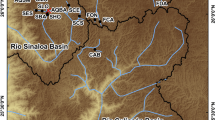
The McCloud River Redband Trout (MRRT; Oncorhynchus mykiss stonei, Jordan 1894) is a subspecies of rainbow trout (O. mykiss ssp.) that has been isolated in the Upper McCloud River for tens of thousands of years by a series of waterfalls that arose through volcanic activities (Fig. 1; Legendre et al. 1972; Miller 1972; Moyle et al. 2008). The long isolation of MRRT is indicated by their unique ancestral characteristics compared to other subspecies of O. mykiss, such as the lowest number of gill rakers, greater number of scales along and above the lateral line, and the frequent presence of vestigial basibranchial teeth (Behnke 1992, 2002). These ancestral characteristics have led to the suggestion that MRRT is a distinctive lineage descended from an early invasion of ancestral trout into the McCloud River headwater system (Behnke 1992, 2002).
Sampling locations of all samples in this study. The left map shows sampling locations of all the samples by watershed, including wild, hatchery rainbow trout, “Other Rainbow Trout”, and Kern River golden trout and rainbow trout. Inset area is the Upper McCloud River (UMCR) watershed. The red box shows sampling locations within the Upper McCloud River watershed, the area above the Middle Falls. Highlighted river in purple is the mainstem McCloud River. Golden Trout Creek (GTCR), South Fork Kern River (SFKR), Kern River (KRNR), Eagle Lake (EGLK), North Fork American River (NFAR), Lower Stanislaus River (LSTN), Lower Yuba River (LYBA), Coleman Hatchery (COLE), Eagle Lake Hatchery (EGLH), Hot Creek Strain (HTCS), Mt. Shasta Hatchery (MTSH), Pit Strain Hatchery (PITS), Warner Valley (WARV), Goose Lake (GOSL), Surprise Valley (SPRV), North Fork Pit River (NFPT), South Fork Pit River (SFPT), Upper Pit River (UPIT), Lower Pit River (LPIT), Yuba North Fork (YUBA), Upper McCloud River (UMCR): Swamp Creek (SWPC), Edson Creek (EDSN), Sheepheaven Creek (SHPN), Dry Creek (DRYC), Moosehead Creek (MOHD), Bull Creek (BLLC), Cow Creek (COWC), Trout Creek (TRTC), Shady Gulch Creek (SHGU), Raccoon Creek (RCCN), McCloud River (MCLD), McKay Creek (MCKY), Blue Heron Creek (BLHN), Tate Creek (TATE)
The Upper McCloud River and MRRT have been heavily impacted by anthropogenic disturbance over the last hundred years, beginning with grazing and logging. Intense cattle grazing in the first half of the 20th century eliminated streamside vegetation, created shallower and wider streams with warmer temperatures, and reduced water quality (Moyle et al. 2008). Logging began in the late 1800s and rapidly expanded with railroad construction through World War II. Logging degraded stream habitat through removal of shade canopy, further increasing in water temperatures, sedimentation, and peak storm flows, while lowering fish habitat diversity (Bolda and Meyers 1997; Moyle et al. 2008). Road construction throughout the Upper McCloud Basin also provided easy access to streams, increasing inputs of sediment and pollutants. However, in the late 20th century, both private and public land managers began to limit logging and grazing in the area (Moyle et al. 2008).
Recurring drought cycles also threaten MRRT, especially given the porous volcanic soil in the region. Even in average or above average water years, many streams in the Upper McCloud region have dry reaches (Pittman 2011). Several times over the past several decades managers have responded to drought conditions and declining MRRT census sizes by translocating fish from drying pools as a conservation measure. Sheepheaven Creek is the most isolated and morphologically distinct population of MRRT (Behnke 2002), and for many years was thought to be the only non-introgressed MRRT population (see below), due to its remote location. In the 1970s, a series of droughts led managers to translocate MRRT from Sheepheaven Creek into presumably fishless Trout and Swamp creeks as a safeguard (Nielsen et al. 1999; Simmons et al. 2010). Sheepheaven Creek experienced another bottleneck during the 1990–1994 drought when the population declined to fewer than 200 individuals (Nielsen et al. 1999). More recently, severe drought conditions from 2013 to 2015 spurred the California Department of Fish and Wildlife (CDFW) to “rescue” MRRT from three populations (Edson, Swamp, and Moosehead creeks) and place them in the Mount Shasta Hatchery to prevent local extirpation (CDFW 2017). These creeks were prioritized for rescue because genetic analyses (Simmons et al. 2010) indicated that these three populations, along with Sheepheaven Creek had not become introgressed (i.e., were genetically “pure” MRRT) with hatchery rainbow trout (O. mykiss ssp.) (see below). The rescued MRRT were spawned across populations in captivity before they could be returned to the wild, founding a new captive MRRT broodstock. Since then, their progeny have been used to stock the Upper McCloud River and McCloud Reservoir with MRRT for recreational angling and overall population reinforcement. As the risk of severe drought increases with climate change, continued management efforts to limit grazing and logging, combined with captive propagation aim to make MRRT more resilient (Moyle et al. 2008).
Another historical and ongoing major threat to MRRT is the introduction of both distantly and closely related nonnative trout species. Brown trout (Salmo trutta) and brook trout (Salvelinus fontinalis) were introduced as early as late 1800s. Brown and brook trout impact MRRT through a combination of factors such as predation, competition, and disease introductions (Fausch et al. 2009; McGrath and Lewis 2007; Moyle et al. 2008). Yet an even greater threat to MRRT was the introduction of closely related O. mykiss ssp.., leading to introgressive hybridization (i.e., incorporation of alleles from one species into the gene complex of another (Anderson 1949)), which can result in loss of both genetic identity and locally adapted alleles (Allendorf and Leary 1988; Wolf et al. 2001; White et al. 2018). Following introgressive hybridization with a conspecific, the phenotype and genotypes of introduced trout tend to become pronounced over time, leading to the erosion of the distinctive characteristics of the native taxon (Wilde and Echelle 1992; Moyle et al. 2008; Seehausen et al. 2008).
Due to increasing recognition of the threat of introgressive hybridization – both from authorized and unauthorized introductions, the stocking of hatchery rainbow trout was discontinued by CDFW in 1994, after Berg (1994) reported introgression in extant MRRT populations (Nielsen et al. 1999; M. Dege, CDFW, pers. comm.). Subsequently, several rounds of genetic studies were conducted to discern levels of introgression in the MRRT populations (Nielsen 1999; Simmons et al. 2010). Nielsen et al. (1999) used microsatellite data and found unique genetic characteristics (e.g., number of alleles, heterozygosity, genetic distance) of Sheepheaven MRRT in comparison to other MRRT. The most recent genetic study was by Simmons et al. (2010), who expanded the sampling effort of Nielsen and used ten SNPs (one mitochondrial and nine nuclear SNPs). Simmons found no evidence of introgression in four out of nine sampled MRRT populations (Sheepheaven, Moosehead, Edson, and Swamp creeks) and only low levels of introgression in several other MRRT populations. Based on Simmons et al. (2010), these four non-introgressed populations are currently classified by CDFW as the “core MRRT conservation populations”, and due to the complex geology and hydrology of the Upper McCloud Watershed, are almost entirely isolated except during very rare high flow events (Nielsen et al. 1999; Moyle et al. 2008). In addition, CDFW wrote a genetic monitoring plan as part of a strategy to manage and protect MRRT, including preventing, detecting, and monitoring introgression from any unauthorized stocking. For MRRT, the threats of small population sizes, hatchery propagation, and unauthorized stocking make ongoing management actions and genetic monitoring necessary; the first priority for managers is to genetically evaluate MRRT core populations.
This project aims to develop genetic tools to aid in monitoring of introgression and genetic diversity in MRRT populations. Specific goals are to: (1) test previous “core MRRT conservation population” designations with genomic data; (2) evaluate additional putatively non-introgressed populations for possible designation as core conservation populations; and (3) develop two sets of genetic markers for rapid genetic monitoring of MRRT: markers that are diagnostic between MRRT and introduced O. mykiss ssp.. to monitor introgression, and markers that are polymorphic within MRRT to monitor genetic diversity (e.g., overall diversity and inbreeding) in both the wild and captivity. Managers can use these markers to identify and adapt to changes in introgression levels and genetic diversity to make informed decisions for conservation actions.
Materials and methods
Sample collection and RAD library preparation
We compiled DNA from 308 individuals from a variety of MRRT and other rainbow trout subspecies in California (Table 1). We included every California rainbow trout subspecies for two reasons. First, the placement of MRRT in phylogenetic analysis has not been consistent among previous studies (Berg 1987; Nielsen et al. 1999). Second, the hatchery stocking and translocation records of rainbow trout into the Upper McCloud River watershed are incomplete and do not include unauthorized introductions (Simmons et al. 2010; M. Dege, CDFW, pers. comm.). The sample collection includes individuals from either archived (some of which were also used in Simmons et al. 2010]) or newly collected samples (Table 1). Our study included three general groups of samples. In addition to the MRRT sample group, we included a group of various rainbow trout samples which we refer to as the “Rainbow Trout Group” (RBTG). The RBTG group includes wild rainbow trout from Surprise Valley, Goose Lake, Pit River, and Warner Valley (also commonly called “redband trout”, but here referred to these as “Other Rainbow Trout” because redband trout does not refer to a monophyletic group). The RBTG group also includes hatchery rainbow trout strains (Coleman, Pit, Hot Creek, Mt. Whitney, Mt. Shasta, and Eagle Lake hatcheries), and wild rainbow trout from North Fork American River, Eagle Lake, and Yuba River. The other sample group is called the “Golden Trout Complex” (GTCX) and includes a representative sample of O. mykiss ssp. fish from Golden Trout Creek, South Fork Kern River, Little Kern River, and Kern River watersheds (Table 1; Fig. 1).
We extracted DNA using the DNeasy extraction kit according to the manufacturer’s protocols (Qiagen). After extraction, we prepared libraries for Restriction Site Associated DNA sequencing (RAD-seq) with the SbfI enzyme based on the protocol described in Ali et al. (2016). For sequencing, all libraries were pooled into a single lane for paired end 150 bp sequencing on an Illumina HiSeq 4000 at UC Davis Genome Center.
Alignment, filtration, and population genetic analysis
After sequencing, we de-multiplexed the data into individual samples and aligned them to the rainbow trout reference genome (Pearse et al. 2019) using the MEM algorithm implemented in the software program BWA (Li and Durbin 2009) to generate Sequence Alignment Map (SAM) files for each individual. The SAM files were then converted to Binary Alignment Map (BAM) files using SAMTOOLS (Li and Durbin 2009; Li 2011). We then used SAMTOOLS to sort, filter for proper pairs, remove PCR duplicates, and index the BAM files. At this stage, 19 individuals were removed due to low read numbers (less than 100,000 mapped reads), with 289 individuals remaining. Then, sequencing coverage was assessed across all discovered loci in each individual, using samtools depth. The mean individual coverage was 9.60, with a maximum of 52.3838, a minimum of 2.00781, and a standard deviation of 7.60 (Fig. S1).We conducted population genetic analyses in ANGSD (Korneliussen et al. 2014), which analyzes BAM files using on a probabilistic framework in the form of genotype likelihoods. For the analyses, we used the SAMTOOLS genotype likelihood model (-GL 1) with a minimum base quality of 10 (-minQ 10) and minimum mapping quality of 20 (-minMapQ 20).
We performed hierarchical population structure analyses: (1) on all samples to identify potential sources of introgression with MRRT, (2) on MRRT and potential introgression sources to estimate introgression levels as accurately as possible, and (3) on just MRRT to examine within-group population structure. For each analysis, we used PCA plots and admixture analyses. For PCAs, we used PCAngsd (Meisner and Albrechtsen 2018) on all samples that passed initial quality filtering to produce a covariance matrix. Our PCAngsd parameters were: SNP_pval = 1e-6, -doMajorMinor, and -doMaf 1. In addition, sites had to have a -minMaf 0.05 and be present in at least 50% of the individuals. We then used admixture analyses to test various population groupings based on genetic structure and shared ancestry. To do this, we used NgsAdmix (Skotte et al. 2013) to estimate admixture proportions of individuals with various cluster (K) values based on genotyped likelihoods calculated in ANGSD (Korneliussen et al. 2014). We conducted 10 runs for each K value, K = 2–6.
To assess genetic diversity of each population, we used theta statistics in ANGSD (-doThetas) and thetaStat (-do_stat) programs (Korneliussen et al. 2013, 2014). To do this, Site Frequency Spectrum (SFS) (-doSaf) was used as a prior to calculate Tajima’s Ɵ (Ɵπ) (Tajima 1983). Tajima’s Ɵ estimates theta (Ɵ = 4Nµ) based on the average number of pairwise nucleotide differences, and when genomic data is used, accurate estimates can be made with even a small sample size (Nelson et al. 2012; Subramanian 2016). Subsequently, two-dimensional SFS (2D-SFS) was applied to calculate pairwise Fst value which represents genetic differentiation between two populations.
Discovery of candidate diagnostic and polymorphic loci for MRRT populations
Using the results from our admixture and PCA analyses, we designed two sets of candidate SNP loci: one to detect levels of introgression between MRRT and other rainbow trout taxa, and a second to monitor within-MRRT genetic diversity. To design the introgression markers (also referred to as MRRT diagnostic markers), we used two overlapping groups of MRRT: MRRTA and MRRTp. MRRTA included all MRRT sampled, even those known to have low levels of introgression, and MRRTp only includes MRRT that are putatively “pure” (i.e., no detectable introgression) by our population structure analyses (the four core populations, Swamp, Sheepheaven, Edson, and Moosehead creeks, plus Dry Creek; see below). We then used these two sample groups to find loci with substantial allele frequency differences between MRRT and a reference rainbow trout (RBT) group (a subset of RBTG) that includes wild and hatchery rainbow trout (Eagle Lake, North Fork American River, Lower Yuba River, Lower Stanislaus River, Coleman Strain, Eagle Lake Strain, Hot Creek Strain, Mt. Shasta Strain, Pit Strain).
We performed genotype calling in ANGSD to find candidate SNPs with alleles fixed or nearly-fixed in MRRTA or MRRTP but not present or present at very low frequency in the RBT group. More specifically, after genotype calling, we calculated the allele frequencies of each locus in each group and selected loci where one allele had a frequency of ≥ 90% in the MRRT group and < 10% in the RBT group. Since MRRTP is not introgressed, we expected a higher number of MRRT-unique markers to be captured compared to MRRTA; therefore, for MRRTP, we increased our cutoff to 99% to discover loci with a higher degree of differentiation (i.e. we selected loci with a frequency of ≥ 99% in the MRRT group and < 1% in the RBT group). In summary, the < 10% filter was used for both MRRTA and MRRTP, and the < 1% filter was used for MRRTP, specifically. Markers specific to MRRT (both groups) are appropriate for monitoring the entire MRRT population including pure and introgressed, and markers specific to MRRTP are appropriate for monitoring pure MRRT populations.
In a separate analysis, we investigated variable SNP loci suitable for monitoring the genetic attributes of both MRRTA and MRRTP groups. We performed genotype calling using the same pipeline applied for introgression markers, except we used a Hardy Weinberg test filter (-doHWE 1) to remove paralogs. Paralogous loci can cause the misidentification of heterozygous/homozygous genotypes. After removing paralogs, we selected polymorphic loci with minor allele frequencies between 0.25 and 0.45 in each of MRRT groups. We applied this range of allele frequencies to capture loci that are polymorphic with a moderate frequency: alleles with a lower frequency (< 0.25) are not informative in many individuals, and alleles with a higher frequency (0.45–0.5) can be duplicates (i.e., paralogous loci) so this filter further reduces paralogs. We performed the process separately for MRRTA and MRRTP with the same allele frequency (between 0.25 and 0.45). Furthermore, to avoid linkage disequilibrium (LD) between selected loci in both polymorphic and diagnostic, we chose loci that are at least 5,000,000 bp apart.
Validation analysis
We conducted a validation analysis to determine whether the MRRTA diagnostic markers identified above were truly diagnostic for the MRRT lineage. We designed SNPtype (Fluidigm Corp.) genotyping assays for a set of 44 candidate SNP loci from MRRTA loci. We obtained dried fin clip samples from fish collected between 2002 and 2007 from three putatively pure MRRT populations (Sheepheaven, Swamp, and Edson creeks) and three populations of hatchery rainbow trout from Crystal Lake (Coleman and Pit strains) and Darrah Springs (Eagle Lake strain) hatcheries. All samples used for validation analyses were taken from the collection of samples used by Simmons et al. (2010) to ensure consistency and allow for comparison across studies. We extracted DNA from the fin clips on the Hamilton Microlab NIMBUS® HD (Hamilton Company) using the Omega Bio-tek Mag-Bind® Blood & Tissue DNA HDQ 96 Kit (Omega Bio-tek, Inc.) according to the manufacturer’s instructions. We then amplified the 44 candidate MRRTA diagnostic SNP loci using Fluidigm® SNP Type Assays and the Juno 96.96 Genotyping Integrated Fluidic Circuit (IFC) on the Juno™ instrument (Fluidigm Corporation) following the manufacturer’s protocol. The Fluidigm SNP genotyping method first uses a locus-specific primer (LSP) and a specific target amplification (STA) primer to enrich for DNA sequences containing the SNP of interest. After enrichment, the LSP and two fluorescently labeled allele specific primers (ASP1 and ASP2) amplify the two possible SNP alleles at a particular locus. We collected fluorescent end-point reads on the Biomark™ HD instrument and used the Fluidigm® SNP Genotyping Analysis Software (version 4.5.1) for SNP allele scoring.
Results
Overall population structure and admixture
The analysis that included all samples identified 284,566 SNPs. Three clear clusters were distinguishable in the PCA that corresponded to the Upper McCloud River redband trout (MRRT), rainbow trout (RBTG), and golden trout (GTCX) groups (Fig. 2: top plot; Table 1). The first principal component (PC1) separates MRRT from GTCX and RBTG, and the second principal component (PC2) separated MRRT from GTCX (Fig. 2: top plot). The linear distribution of MRRT along PC1 towards RBTG suggests introgression with this group rather than GTCX. The results in the PCA are supported by the admixture analyses with all samples included, using K = 2 and K = 3 (Fig. 2: bottom plot; see Fig. S2 and Table S1 for the higher K plots and the likelihood for each K). At K = 2, MRRT is differentiated from RBTG and GTCX, and at K = 3, GTCX, RBTG, and MRRT are three distinct clusters, but there is about 20–25% RBTG ancestry within much of the MRRT group. After confirming that all of our Upper McCloud River samples clustered together and were genetically separated from the other trout subspecies, we next attempted to distinguish putatively pure and introgressed populations of MRRT. From the previous PCA and admixture result (Fig. 2), it is clear that MRRT are an independent cluster that does not group with GTCX or RBTG. However, the MRRT distribution on PC1 trends towards a subset of RBTG (a small cluster at the right end of the RBTG main cluster), which suggests that introgression in MRRT is with a subset of RBTG (Fig. 2). This RBTG subset cluster includes wild rainbow trout from Eagle Lake and the American River, Steelhead trout from the Yuba and Stanislaus Rivers, hatchery strains (Shasta, Coleman, Eagle Lake, Hot Creek strains) and three creeks from the “Other Rainbow Trout” group: Lincoln, Lost, and Nelson creeks (Table 1). Hence, we performed PCA and admixture analyses on MRRT and this subset of RBTG to most accurately identify the introgression level in MRRT populations (Fig. 3).
Population Structure of all samples. Top plot: all samples PCA, color represents watershed. Three main groups are distinguishable: Golden Trout Complex (GTCX), Rainbow Trout Group (RBTG), and McCloud River Redband Trout (MRRT). PC1(8.7% variance explained) / PC2(7.65% variance explained). Bottom plot: all samples admixture plots at K = 2 (top admixture plot) and K = 3 (bottom admixture plot). Blue represents MRRT ancestry group which is different from GTCX (green) and RBTG (red). Golden Trout Creek (GTCR), South Fork Kern River (SFKR), Kern River (KRNR), Eagle Lake (EGLK), North Fork American River (NFAR), Lower Stanislaus River (LSTN), Lower Yuba River (LYBA), Coleman Hatchery (COLE), Eagle Lake Hatchery (EGLH), Hot Creek Strain (HTCS), Mt. Shasta Hatchery (MTSH), Pit Strain Hatchery (PITS), Warner Valley (WARV), Goose Lake (GOSL), Surprise Valley (SPRV), North Fork Pit River (NFPT), South Fork Pit River (SFPT), Upper Pit River (UPIT), Lower Pit River (LPIT), Yuba North Fork (YUBA), Upper McCloud River (UMCR)
PCA and admixture analyses of MRRT with a group of a potential source of introgression – a small subset of RBTG. Top plot: PCA of MRRT with the RBTG small subset, color represents populations. The RBTG small subset includes: wild rainbow trout from Eagle lake (EGLK) and North Fork American River (NFAR), Steelhead from Lower Yuba River (LYBA) and Stanislaus River (LSTN), hatchery strains from Mt. Shasta (MTSH), Coleman (COLE), Eagle Lake (EGLH), Hot Creek (HTCS), and three from the “Other Rainbow Trout” group: Lincoln (LCLN) and Lost (LOST) from Lower Pit River watershed and Nelson (NLSN) creeks from Yuba watershed, PC1(11.6% variance explained) / PC2(3.44%, variance explained). Bottom plot: admixture plot of MRRT and the RBTG subset cluster. Five pure populations are identified within the MRRT population: Swamp creek (SWPC), Edson Creek (EDSN), Sheepheaven Creek (SHPN), Dry Creek (DRYC), and Moosehead Creek (MOHD)
The PCA performed with MRRT and the RBTG subset cluster (Fig. 3) suggests that hatchery and wild Eagle Lake rainbow trout populations, which are clustered separately from the rest of the RBTG subset (Fig. 3: top plot, top right populations in yellow and red), are not the likely source of introgression. In addition, the MRRT individuals’ linear distribution along the PC1 towards the RBTG cluster suggests introgression of some MRRT populations (Fig. 3: top plot). An admixture analysis on the same MRRT and RBTG subset sample groups strongly supports the PCA result (Fig. 3: bottom plot). To quantify this, we calculated the correlation coefficient between PC1 value and the proportion of rainbow trout ancestry (%RBT) of each individual. The correlation coefficient of 99% (r = 0.995) demonstrates that MRRT individuals/populations with higher %RBT have higher PC1 values (Table S2).
Our admixture plot showed no evidence of introgression in individuals tested in five populations from the Upper McCloud watershed: Swamp, Edson, Sheepheaven, Dry, and Moosehead creeks (Fig. 3: bottom plot). However, there is apparent introgression in individuals from Trout, Blue Heron, Raccoon, Cow, Bull, Tate, McKay, Shady Gulch creeks. Furthermore, six sampling locations in the mainstem Upper McCloud River show varying levels of introgression (Fig. 3: bottom plot). The levels of introgression vary among the locations (Table 2). For example, Bull, Cow, and Trout creeks have the lowest introgression levels (mean %RBT of 0.0205, 0.0547, and 0.0980, respectively), with other locations showing higher levels of introgression (> 0.1910).
Within MRRT population structure
To assist managers in prioritizing MRRT populations for further population genetic investigation, we examined population structure within MRRT. To do this, we conducted admixture analyses with only the MRRT group at K = 2–5 (Fig. S3). Considering five identified pure MRRT populations, and the fact that at least one of the populations is founded by another (Swamp Creek by Sheepheaven Creek), we assessed K = 3 and 4 for the population structure interpretation (Fig. 4; Table S1). We found three main genetic groups (this excludes the rainbow trout genetic group represented in green) at K = 4: red representing Swamp and Sheepheaven creeks genetic group, blue representing Edson and Dry creeks genetic group, and purple representing Bull Creek genetic group (Fig. 4, K = 4).
Population structure within MRRT group. MRRT group admixture plot at K = 3 and K = 4. Four major genetic groups are distinguishable: red represents Swamp and Sheepheaven creeks genetic group, blue represents Edson and Dry creeks genetic group, purple represent Bull creek genetic group, and green represents rainbow trout genetic group. The population’s order is based on increasing in the percentage of rainbow trout ancestry.
Swamp Creek (SWPC), Edson Creek (EDSN), Sheepheaven Creek (SHPN), Dry Creek (DRYC), Moosehead Creek (MOHD), Bull Creek (BLLC), Cow Creek (COWC), Trout Creek (TRTC), Shady Gulch Creek (SHGU), Raccoon Creek (RCCN), McCloud River (MCLD), McKay Creek (MCKY), Blue Heron Creek (BLHN), Tate Creek (TATE)
We calculated Tajima’s theta (Ɵπ) (Table 3) to quantify the overall genetic diversity of the pure MRRT populations. Dry and Edson had the highest (0.000699) and lowest (0.000474) Ɵπ values, respectively, although Moosehead Creek’s Ɵπ value (0.000691) is almost as high as Dry Creek’s. Sheepheaven and Swamp creeks had intermediate Ɵπ values (0.000592 and 0.000561, respectively). We also measured genetic differentiation between pure populations using pairwise Fst (Table 3). Since Moosehead Creek’s sample size was different from the other four populations (2 individuals vs. 5 individuals), we did not consider any pairwise Fst with the Moosehead Creek in the Fst result. However, among the rest of the pairwise comparisons, Swamp vs. Sheepheaven had the lowest (0.1061) and Edson vs. Swamp had the highest Fst value (0.3989 ).
Diagnostic loci for MRRT
We identified putatively non-introgressed MRRT populations based on our admixture and PCA results and identified individuals for two MRRT groups (MRRTA and MRRTP). We used N = 57 individuals from the MRRTA population group and N = 22 for the MRRTP group (Sheepheaven, Edson, Swamp, Moosehead, and Dry creeks). The RBT group contained 35 wild and hatchery rainbow trout individuals (Eagle Lake, North Fork American River, Lower Yuba River, Lower Stanislaus River, Coleman Strain, Eagle Lake Strain, Hot Creek Strain, Mt. Shasta Strain, Pit Strain).
We discovered 44 differentially fixed SNPs between the broader MRRTA group and RBT. We found 2,649 diagnostic SNPs at a frequency of ≥ 90% between the MRRTP and RBT (Table 4). When we increased the minimum frequency of diagnostic alleles to 99% in the MRRTP, the number of SNPs was reduced to 574. All of these 574 fixed SNPs had the frequency of 100% in MRRTP and 0% in RBT before validation (Table 4). Of the MRRTA loci, 80% were found in MRRTP with the 90% cutoff, and 50% were found in MRRTP with the 99% cutoff. For polymorphic markers, we found 6,639 loci in MRRTA and 7,316 in MRRTP (Table 4).
Validation results
We successfully amplified and visualized callable heterozygote and homozygote clusters in 27 of the 44 candidate MRRTA diagnostic SNP loci using the Fluidigm SNP Type assays (Tables S3 and S4). There were two loci (omy01_36537055 and omy15_57867903) that failed to amplify in > 90% of individuals in the MRRTP and the tested hatchery rainbow trout populations; this data should therefore be interpreted with caution (denoted by asterisks in Table 5). We obtained genotypes for 90 MRRT from putatively non-introgressed locations and 87 hatchery rainbow trout individuals for at least 90% of these two loci; we discarded data from one individual from Edson Creek because it did not meet this threshold. Of the 27 remaining MRRTA diagnostic loci, 16 were completely fixed between the groups of MRRTA and hatchery rainbow trout that we tested (Table 5). The frequency of MRRT alleles was greater than 95% for all historically non-introgressed MRRT populations that we tested (Swamp, Sheepheaven, and Edson creeks) in 21 of the candidate diagnostic loci (Table 5); we propose that these loci should be used in assessing introgression between MRRTA and MRRTP with hatchery rainbow trout as part of future monitoring efforts.
Discussion
Using population structure analysis, we identified potential MRRT introgression sources from RBTG and we confirmed former findings of four non-introgressed MRRT locations (Edson, Swamp, Sheepheaven, Moosehead), and potentially one more (Dry). In addition, we identified three main genetic groups with MRRT: Sheepheaven and Swamp creeks, Edson and Dry creeks, and Bull Creek. Using these results, we discovered and validated diagnostic and polymorphic SNP loci specific to the MRRTP (Swamp, Edson, Sheepheaven, Moosehead, and Dry creeks) and MRRTA (Swamp, Edson, Sheepheaven, Dry, Moosehead, Blue Heron, Bull, Cow, Tate, McCloud River, Raccoon, Trout, McKay, and Shady Gulch Creek) groups to monitor their introgression and genetic diversity both in the wild and the captive breeding program.
Ancestral history of MRRT
Previous research has identified morphological similarities (Schreck and Behnke 1971; Hoopaugh 1974; Gold 1977) and a shared karyotype (2n = 58; Thorgaard 1983) between GTCX and MRRT. Behnke (1981) reconciled their morphological similarities and in light of their geographic separation by suggesting that GTCX may have resulted from multiple invasions of a primitive redband trout via the Sacramento River and Tulare Lake, and that all ancient redbands were subsequently extirpated except for those in the Upper McCloud River. However, Gall et al. (1981) found that the trout in the Upper McCloud River differ substantially from golden trout and rainbow trout by meristic, chromosomal, and electrophoretic characteristic traits. Furthermore, subsequent microsatellite data found that the golden trout complex (except Little Kern golden trout) is significantly different from the MRRT (Nielsen et al. 1999), but the authors suggested that inbreeding and introgression could cause significant genetic difference when two stocks with a common ancestry are isolated. Later, Stephens (2007) also reported a more distant common ancestor of MRRT and golden trout complex from AFLP data.
In our analysis we found three main groups (MRRT, RBTG, and GTCX) using PCA and admixture analyses in our samples (Fig. 2). Despite the fact that the level of introgression in the MRRT group found by the admixture analysis was not very high, at K = 2, the introgression source in MRRT appears to be from the RBTG and GTCX ancestry group because these two groups are combined at K = 2. However, at K = 3, when GTCX and RBTG are split, the introgression source is RBTG (Fig. 2: bottom plot). Our assessment that MRRT is separate from both RBTG and GTCX is in line with Behnke’s (1992) claim that MRRT should have subspecific status. Thus, we did not find evidence of recent shared ancestry between MRRT and the golden trout complex.
Potential sources of introgression
Most hatchery strains of rainbow trout (primarily O. m. irideus) used in California were derived from rainbow trout in the upper Sacramento River (Nielsen et al. 1999). The Eagle Lake rainbow trout (O. m. aquilarum) was also stocked extensively in California for several decades and is still in use by CDFW. Given the long history of trout stocking (hatchery records indicating stocking of Mt. Shasta hatchery trout in the Upper and Lower McCloud River (M. Dege, CDFW, pers. Comm.)) and the geographic proximity to the McCloud River, we expected the Mt. Shasta Hatchery (Mt. Shasta, California) to be the most probable source of planted fish in the McCloud River. Prior to Mt. Shasta Hatchery, there was also extensive stocking of trout beginning in the late 1800s from Baird Hatchery, which was located near the confluence of the McCloud River and the Pit River but covered by Lake Shasta following the construction of Shasta Dam in the 1940s. Indeed, Berg (1994) reported hatchery introgression of the Upper McCloud fish by assessing several protein loci, ultimately leading to the end of the hatchery stocking in 1994.
Our results show that only a small subset of the rainbow trout that we examined (Fig. 2: top plot, bottom right of top left cluster) can be the most likely source of introgression. This small group includes wild and hatchery rainbow trout, and three populations from the “Other Rainbow Trout” group. Among the hatchery strains used in this study, Mt. Shasta, Coleman, and Hot Creek represent potential sources of introgression, but Pit Strain and possibly Eagle Lake strain do not. Pit Strain is markedly different from other hatchery trout strains used in California; Pit strain rainbow trout were originally sourced from the Pit River, which falls within the geographic range of the general redband trout designation. It is notable that Nielsen et al. (1999) assessed the same hatchery groups as our study but only found significant genetic association (low genetic distance, RST) between Sheepheaven Creek MRRT and Eagle Lake hatchery strain rainbow trout, but not other hatchery strains. The conflict may be explained by the power of RAD sequencing with a much greater coverage and thousands of markers in comparison to highly variable but limited number of microsatellite markers for discovering and genotyping polymorphic loci. In addition, mainstem Upper McCloud River has been extensively stocked with a variety of rainbow trout strains and individual hatcheries often stock multiple different strains of domesticated rainbow trout. For example, Mt. Shasta Hatchery has used Shasta, Coleman and Eagle Lake strain trout for various stocking activities (M. Dege, CDFW, pers. comm.). Additionally, incomplete records of stocking from the past century and ad hoc crosses between hatchery strains can explain observing multiple hatchery strains as a source of introgression. Identifying this small group as the source of introgression allows us to more accurately quantify introgression level.
Levels of Introgression
Our results are generally consistent with Nielsen et al. (1999) and Simmons et al. (2010) in supporting the classification of Edson, Swamp, Sheepheaven, and Moosehead creeks as non-introgressed MRRT core conservation streams. As part of marker discovery, we also included samples from five other creeks in the Upper McCloud watershed: Dry, Bull, Cow, Shady Gulch, and Blue Heron. We only used 3–5 samples from each of these creeks for MRRT diagnostic marker discovery, but our introgression analyses suggest that further genetic investigation of Dry Creek as a candidate core conservation stream is warranted because we detected no rainbow trout introgression in our samples. We also found only minor admixture influence in Bull and Cow creeks and recommend that additional genetic sampling be performed to discern whether these creeks should be considered core conservation streams. Furthermore, in future analyses it is important to include samples from different reaches of these creeks; introgression is believed to vary with hydrological connectivity, accessibility, and the potential for unauthorized stocking (M. Dege, CDFW, pers. comm.).
The unique hydrology and connectivity patterns in the Upper McCloud River could explain the introgression patterns we observed. In general, we expect lower stream connectivity and isolation from the public to correlate with lower levels of introgression. The northern streams (Swamp, Edson, Sheepheaven, and Trout creeks) are the least introgressed and the most isolated. These locations are mostly disconnected from the mainstem Upper McCloud except during rare high-flow events (Nielsen et al. 1999) and are largely on private timber properties (M. Dege, CDFW, pers. comm.). The only non-introgressed population in the south is Moosehead Creek. Although Moosehead Creek is regularly connected by surface flows, a concrete fish barrier near its confluence with the Upper McCloud River prevents fish from ascending into the creek (M. Dege, CDFW, pers. comm.). The tributaries that are mainly south of the Upper McCloud River are expected to be introgressed, because they are connected to the mainstem Upper McCloud River and may have been stocked with hatchery rainbow trout in the past. For example, Raccoon and Tate creeks showed introgression consistent with Simmons et al. (2010). Raccoon and Tate creeks lack fish barriers between the mainstem Upper McCloud River and are easily accessible to the general public, which may explain the observed higher levels introgression.
We expected that Swamp and Trout creeks, which were recipients of MRRT from Sheepheaven Creek in the 1970s, to be genetically similar to Sheepheaven Creek. However, in the admixture analysis, Trout Creek shows low levels of introgression (0.098), but Swamp does not (Fig. 3). Prior to stocking MRRT, there was a rainbow trout eradication action in Trout Creek which may have been incomplete, or rainbow trout may have been unlawfully introduced via a public campground in the Lower Trout Creek. However, Pittman (2011) reported MRRT as the only salmonid species observed in Trout Creek. Although Trout Creek has no discernable barriers to fish passage, the levels of introgression are highly dependent on sampling location, with lower Trout Creek showing patterns of introgression while upper Trout Creek appears to be more “pure” MRRT (Stephens et al. 2013). In addition to Trout Creek, Bull and Cow creeks had low levels of introgression (0.0205, 0.0547, and 0.098 respectively). These creeks are hydrologically isolated from Upper McCloud River most of the year but may be connected during seasonal or higher flow events, especially in the lower section of Bull Creek (S. Plemons and M. Dege, CDFW, pers. comm.) leading to possible introgression with RBT.
Pure MRRT have three genetic groups
We observed three genetic groups within pure MRRT using admixture analyses: (1) Sheepheaven and Swamp creeks, (2) Edson and Dry creeks, and (3) Bull Creek (Fig. 4, K = 4). Overall, our Fst results support the observed genetic clusters so that the genetic differentiation between the two genetic clusters (Edson and Dry creeks’ pairwise Fst with Swamp and Sheepheaven creeks, Table 3) are higher than within clusters’ pairwise Fst (Edson Creek vs. Dry Creek and Swamp Creek vs. Sheepheaven Creek, Table 3). Except, Dry vs. Edson creeks’ Fst (0.3262) that is higher than Dry vs. Sheepheaven creeks’ (0.3005), which may be explained by the lower genetic diversity in Edson caused by genetic drift. Sheepheaven and Swamp creeks’ common ancestry on the admixture plots is consistent with the fact that Swamp Creek was historically fishless prior to a translocation of MRRT from Sheepheaven Creek. Similar to Simmons et al. (2013), our genetic diversity analysis shows that Swamp Creek has a slightly lower genetic diversity (Ɵπ = 0.000561) than Sheepheaven (Ɵπ = 0.000592) which is expected for a derived population and consistent with founder effects (Table 3); they also have the lowest pairwise Fst value (0.1061, Table 3). Similarly, Trout Creek was founded by a translocation from Sheepheaven Creek and shows a higher proportion of common ancestry with Sheepheaven and Swamp creeks than with other introgressed populations.
Interestingly, the Edson and Dry creek genetic group was present in most MRRT populations. Thus, this genetic group appears to be the most common historical genetic group within MRRT populations rather than the expected Sheepheaven Creek genetic group (Fig. 4). Sheepheaven was previously thought to be the “sole representative” of MRRT (Behnke 1992, 2002). Simmons et al. (2013) 2010also found private alleles specific to Edson and Moosehead which were not present in Sheepheaven. For the same reason, it is also surprising that Bull Creek, which has the same genetic group as Edson, Dry, and Moosehead at K = 3, shows a unique MRRT signature at K = 4.
Our results have significant implications for the management of MRRT. For example, our results suggest that Sheepheaven Creek is not the only representative of ancestral MRRT lineages. Edson and Dry creeks may also represent a distinct genetic group. In addition, our analyses show that Edson Creek has the lowest genetic diversity (Ɵπ = 0.000474). This may imply that Edson Creek can benefit from targeted conservation management such as supplementation from other pure MRRT populations either within the same genetic group (e.g. Moosehead) if managers want to preserve the genetic group or from another group if they want to increase overall genetic diversity. However, Simmons et al. (2013) reported low allelic richness for Edson and Sheepheaven, and a bottleneck in Sheepheaven based on microsatellite data. Hence, we need to take into account the possibility that the observed clusters might have appeared due to small population sizes experiencing genetic drift instead of representing distinct historical lineages. Therefore, additional genetic testing with a broader sampling distribution within each tributary is warranted to further refine MRRT conservation efforts for each individual creek. Furthermore, to prioritize the populations for conservation purpose based on their adaptive potential, neutral genetic diversity is not enough and other factors must be considered such as balancing selection on particular segregating sites, effective population size, rate of mutation, and the populations’ adaptive diversity (Messer and Petrov 2013; Kardos and Luikart 2021; Teixeira and Huber 2021).
Diagnostic and polymorphic markers
We identified SNP loci that are both diagnostic for identifying non-introgressed and introgressed MRRT and loci that are polymorphic in MRRT. The development of new SNP type assays will facilitate rapid, consistent genetic typing of individual fish from both pure MRRT and introgressed populations to help inform adaptive conservation strategies and actions. We found 44 diagnostic loci (21 after validation) for the entire MRRT population and more than 500 SNP loci for the non-introgressed MRRT, reflective of their unique, divergent lineage from RBTG. Although only MRRTA diagnostic markers were validated, we would expect a similar validation efficiency for the MRRTP markers since the marker discovery methods used for MRRTP and MRRTA were similar.
Management implications and conclusion
The major goal of genetic monitoring in fish and wildlife populations is to help maintain adaptive capacity in unique genetic lineages, providing populations with resiliency against environmental changes into the future. Introgression between threatened native and introduced populations can alter the genetic diversity of native populations and impact their long-term viability (White et al. 2018; Frankham et al. 2009). The historical and ongoing potential for unauthorized introduction of other rainbow trout has exposed the naturally isolated, genetically unique MRRT to introgressive hybridization with nonnative rainbow trout subspecies. Furthermore, MRRT is especially prone to further loss of genetic diversity through processes inherent to small, isolated populations, such as genetic drift and an accumulation of inbreeding effects. Currently, MRRT only exists in a handful of small, isolated populations and also in a conservation hatchery program as a captive broodstock, thus making an adaptive management strategy a necessity.
Admixture, population structure, and genetic diversity analyses conducted in this study corroborate previous work that showed that Edson, Swamp, Sheepheaven, and Moosehead creeks are not introgressed with hatchery rainbow trout. While we found three genetic clusters, this represent different management options in the case of detected inbreeding in populations and the necessity of genetic rescue. Beyond the five populations identified as pure MRRT, we suggest further investigation of Dry, Bull, Trout, and Cow creeks with larger sample sizes to further characterize patterns of introgression and to determine which, if any should be considered for core conservation stream status.
The monitoring and management of introgressive hybridization in MRRT core conservation populations should be a critical objective of an MRRT management strategy. Currently, most of the recent hatchery-produced MRRT have been stocked in the Upper McCloud River. Hatchery-produced F1 juveniles, which were the result of the spawning of rescued wild adult MRRT were stocked into both Trout and Moosehead creeks in a one-time event to help restart the populations following the extreme drought conditions. Currently, no authorized stocking is occurring in the creeks with pure MRRT populations, and further stocking activities will likely be limited to the Upper McCloud River between Algoma (Upper McCloud River) and McCloud Reservoir (M. Dege, CDFW, pers. comm.). Overall, the results from this study will be a valuable resource to assist with future genetic evaluation and monitoring of MRRT.
Availability of data and material:
All sequence data generated in this study are available at the NCBI Sequence Read Archive with identifier SUB9488841. Author contributions: Resources: E.H., M.R.M., J.R., G.M.A., M.S., and A.J.F.; investigation: E.H., M.R.M., and A.J.F.; formal analysis: E.H., M.R.M., and A.J.F; validation: D.G., L.S., and J.R.; visualization: E.H., M.R.M., and A.J.F; writing of original draft: E.H., M.R.M., and A.J.F.; writing, review, and editing: E.H., M.R.M., D.G., L.S., J.R., G.M.A., M.S., and A.J.F.; funding acquisition: M.R.M., J.R., and A.J.F ; supervision: M.R.M., and A.J.F.
Code Availability
Not applicable.
References
Anderson E (1949) Introgressive hybridization. New York, J. Wiley
Ali OA, O’Rourke SM, Amish SJ, Meek MH, Luikart G, Jeffres C, Miller MR (2016) Rad capture (Rapture): Flexible and efficient sequence-based genotyping. Genetics 202(2):389–384
Allendorf FW, Leary RF (1988) Conservation and distribution of genetic variation in a polytypic species, the cut-throat trout. Conserv Biol 2:170–184
Behnke RJ (1992) Native trout of western North America. American Fisheries Society Monograph 6, Bethesda, Maryland
Behnke RJ (2002) Trout and salmon of North America. Free Press, New York
Berg WJ (1987) Evolutionary genetics of rainbow trout, Parasalmo gairdnerii (Richardson). Doctoral dissertation. University of California, Davis
Berg WJ (1994) Introgressive hybridization of McCloud River redband trout by hatchery rainbow trout. Report submitted to California Department of Fish and Game, Wild Trout Division, Sacramento, CA
Bolda KS, Meyers WJ (1997) Conducting a long-term water quality monitoring project: a case study on the McCloud River, California. J Soil Water Conserv 52(1):49–54
CDFW (2017) McCloud Redband post outplanting monitoring field report; California Department of Fish and Wildlife, Northern Region. 7 pp
Gold JR (1977) Systematics of western North American trout (Salmo), with notes on the redband trout of Sheepheaven Creek, California. Calif J Zool 55:1858–1873
Gall G, Bannon M, Smith R, Bentley B (1981) Progress Report. Native Trout of California. California Department of Fish and Game, Threatened Trout Committee. 46 pp
Hoopaugh DA (1974) Status of the Redband Trout (Salmo sp.) in California. California Department of Fish and Game, Inland Fisheries Div. Admin Rpt 74(7):11
Fausch KD, Rieman B, Dunham JB, Young MK, Peterson DP (2009) Invasion versus isolation: trade-offs in managing native salmonids with barriers to upstream movement. Conserv Biol 23:859–870
Frankham R, Ballou JD, Briscoe DA(2009) Introduction to conservation genetics, 2nd edition. Cambridge University Press
Kardos M, Luikart G (2021) The genetic architecture of fitness drives population viability during rapid environmental change. Am Nat 197(5):511–525
Korneliussen TS, Albrechtsen A, Nielsen R (2014) ANGSD: Analysis of Next Generation Sequencing Data. BMC Bioinformatics 15(1):1–13
Korneliussen TS, Moltke I, Albrechtsen A et al (2013) Calculation of Tajima’s D and other neutrality test statistics from low depth next-generation sequencing data. BMC Bioinformatics 14(1):1–14
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Li H (2011) A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics 27(21):2987–2993
Legendre P, Schreck CB, Behnke RJ (1972) Taximetric analysis of selected groups of western North American Salmo with respect to phylogenetic divergences. Syst Zool 21:292–307
McGrath CC, Lewis WM Jr (2007) Competition and predation as mechanisms for displacement of greenback cutthroat trout by brook trout. Trans Am Fish Soc 136:1381–1392
Meisner J, Albrechtsen A (2018) Inferring Population Structure and Admixture Proportions in Low Depth NGS Data. Genetics 210(2):719–731
Meffe GK (1986) Conservation Genetics and the Management of Endangered Fishes. Fisheries, 11(1):14–23
Messer PW, Petrov DA (2013) Population genomics of rapid adaptation by soft selective sweeps. Trends Ecol Evol 28:659–669
Miller RR(1972) Classification of the native trouts of Arizona with the description of a new species, Salmo apache. Copeia, 1972: 401–422
Moyle P, Israel JA, Purdy SE (2008) Salmon, Steelhead, and Trout in California. Status of an Emblematic
Fauna. Center for Watershed Sciences,University of California, Davis
Nelson MR, Wegmann D, Ehm MG, Kessner D, St Jean P, Verzilli C et al (2012) An abundance of rare functional variants in 202 drug target genes sequenced in 14,002 people. Science 337(6090):100–104
Nielsen JL, Crow KD, Fountain MC (1999) Microsatellite diversity and conservation of a relic trout population: McCloud River redband trout. Mol Ecol 8(1):S129–S142
Pittman RG(2011) Minimum Stream Length Requirements for McCloud River Redband Trout (Oncorhynchus mykiss spp.) in Trout and Tate creeks in Siskiyou County, California. Dissertation, Humboldt State University
Pearse DE, Barson NJ, Nome T, Gao G, Campbell MA, Abadía-Cardoso A, Anderson EC, Rundio DE, Williams TH, Naish KA, Moen T, Liu S, Kent M, Moser M, Minkley DR, Rondeau EB, Brieuc MSO, Sandve SR, Miller MR, Cedillo L, Baruch K, Hernandez AG, Ben-Zvi G, Shem-Tov D, Barad O, Kuzishchin K, Garza JC, Lindley ST, Koop BF, Thorgaard GH, Palti Y, Lien S (2019) Sex-dependent dominance maintains migration supergene in rainbow trout. Nat Ecol Evol 3:1731–1742
Ryman N (1991) Conservation genetics considerations in fishery management. Journal of’ Fish Biology, 39:211–224
Schreck CB, Behnke RJ (1971) Trouts of the upper Kern River Basin, California with reference to systematics and evolution of western North American Salmo. J Fish Res Board Can 28:987–998
Skotte L, Korneliussen TS, Albrechtsen A (2013) Estimating individual admixture proportions from next generation sequencing data. Genetics 195(3):693–702
Simmons RE, Lavretsky P, May B (2010) Introgressive Hybridization of Redband Trout in the Upper McCloud River Watershed. Trans Am Fish Soc 139:201–213
Simmons R, Stephens M, May B (2013) Genetic analysis of California redband trout. Final Report to California Department of Fish and Wildlife. CDFW Agreement #P1181006. May 20, 2013. Genomic Variation Laboratory, University of California, Davis
Small MP, Beachem TD, Withler RE, Nelson RJ (1998) Discriminating coho salmon (Oncorhynchus kisutch) populations within Fraser River, British Columbia, using microsatellite DNA markers. Mol Ecol 7(2):141–155
Stephens MR(2007) Contribution of population genetic structure, hybridization, and cultural attitudes to the conservation of threatened native trout. PhD Dissertation. Davis, University of California
Stephens MR, Erickson B, Finger M, Simmons R, May B (2013) Final report. Genetic analysis of Native California Trout (Phase 6). California Department of Fish and Wildlife Agreement # P1181006
Subramanian S (2016) The effects of sample size on population genomic analyses implications for the tests of neutrality. BMC Genomics 17(1):1–13
Tajima F (1983) Evolutionary relationship of DNA sequences in finite populations. Genetics 105:437–460
Teixeira JT, Huber CD(2021) The inflated significance of neutral genetic diversity in conservation genetics
Proceedings of the National Academy of Sciences, 118 (10)
White SL, Miller WL, Dowell SA, Bartron ML, Wagner T (2018) Limited hatchery introgression into wild brook trout (Salvelinus fontinalis) populations despite reoccurring stocking. Evol Appl 11:1567–1581
Wolf DE, Takebayashi N, Rieseberg LH (2001) Predicting the risk of extinction through hybridization. Conserv Biol 15(4):1039–1053
Acknowledgements
The authors would like to thank California Department of Fish and Wildlife (CDFW) for funding the project under contract P1781006. We like to thank all members at CDFW, particularly, Dave Lentz who obtained the funding for this study, Michael Dege, Andrew Jensen, and Samuel Plemons who provided the samples for this project and their communications and information, and Kristen Ahrens who performed genotyping and troubleshooting for SNP assay validation. The authors would like to thank UC Davis Genome Center for data sequencing. We also gratefully appreciate Marin Rod & Gun Club (MR&GC) for their scholarship and support for E H. We would like to thank Matthew A. Campbell, Visiting Scholar at UC Davis Genomic Variation lab, and Ryan Peek, Postdoctoral Scholar at UC Davis Center of Watershed Science, for their inputs.
Funding
This study was funded by California Department of Fish and Wildlife (CDFW) under contract P1781006.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest/Competing interests
We have no conflicts of interest to disclose.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Habibi, E., Miller, M.R., Gille, D. et al. Genomics facilitates evaluation and monitoring of McCloud River Redband Trout (Oncorhynchus mykiss stonei). Conserv Genet 23, 773–789 (2022). https://doi.org/10.1007/s10592-022-01453-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-022-01453-7