Abstract
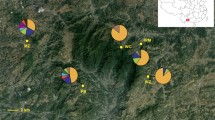
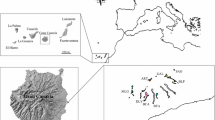
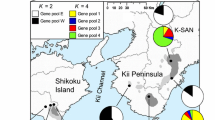
How genetic diversity is maintained within a range of species is one of the crucial pieces of information in species conservation. Although the classical central-marginal hypothesis assumes genetic paucity in peripheral populations, this pattern is not always the case. The semi-shrub Chimaphila umbellata subsp. umbellate, population of which are located in the southernmost part of the subspecies distribution, is considered to be a threatened species in Japan. Thus, this study aimed to examine which populations should be preferentially conserved and if the central-marginal hypothesis can be applied to this case. Genetic diversity was examined in 15 populations of C. umbellata subsp. umbellata in Japan using 16 nuclear simple sequence repeat markers. Overall, the genetic diversity values within the populations did not correlate with the latitude of the population locality, although those of the southern marginal populations tended to be lower than those of populations in other regions. While the populations are genetically differentiated from each other, recent population size declines were only detected in a few cases. Bayesian inference of population structure revealed three genetically distinct groups. An approximate Bayesian computation revealed that these three genetic groups were derived from the ancient population in a recent times. The contribution of each population to total genetic diversity was estimated by removing a given population and recalculating the total genetic diversity. Only one population contributed to both gene diversity and allelic diversity, while several populations contributed to one or the other. Considering genetic diversity and structure, the conservation priority of Japanese populations of C. umbellata to preserve genetic diversity is discussed.





Similar content being viewed by others
Data availability
Gene frequency data of SSR used in the study were given in Online Resource 2.
References
Allendorf FW (2017) Genetics and the conservation of natural populatins; allozymes to genomes. Mol Ecol 26:420–430
Arnaud-Haond S, Belkhir K (2007) GENCLONE: a computer program to analyse genotypic data, test for clonality and describe spatial clonal organization. Mol Ecol Notes 7:15–17
Assis J, Coelho NC, Alberto F, Valero M, Raimondi P, Reed D, Serrao EA (2013) High and distinct range-edge genetic diversity despite local bottlenecks. PLoS ONE 8:e68646
Bai WN, Liao WJ, Zhang DY (2010) Nuclear and chloroplast DNA phylogeography reveal two refugium areas with asymmetrical gene flow in a temperature walnut tree from East Asia. New Phytol 188:892–901
Barrett SCH, Kohn JR (1991) Genetic and evolutionary consequences of small population size in plants: implications for conservation. In: Falk DA, Holsinger KE (eds) Genetics and conservation of rare plants. Oxford University Press, New York, pp 3–30
Bertorelle C, Benazzo A, Mona S (2010) ABC as a flexible framework to estimate demography over space and time: some cons, many pros. Mol Ecol 19:2609–2625
Blacket MJ, Robin C, Good R, Lee SF, Miller AD (2012) Universal primers for fluorescent labeling of PCR fragments: an efficient and cost-effective approach to genotyping by fluorescence. Mol Ecol Resour 12:456–463
Booy GR, Hendriks RJJ, Smulders MJM, Van Groenendael J (2000) Genetic diversity and the survival of populations. Pl Biol 2:379–395
Brownstein MJ, Carpten JD, Smith JR (1996) Modulation of non-templated nucleotide addition by Taq DNA polymerase: primer modification that facilitate genotyping. Biotechniques 20:1004–1010
Carbognani M, Piotti A, Leonardi S, Pasini L, Spanu I, Venderamin GG, Tomaselli M, Petraglia A (2019) Reproductive and genetic consequences of extreme isolation in Salix herbacea L. at the rear edge of its distribution. Ann Bot 124:849–860
Chapuis MP, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Comes HP, Kadereit JW (1998) The effect of Quaternary climate changes on plant distribution and evolution. Trends Plant Sci 3:432–438
Cornuet JM, Ravigné V, Estoup A (2010) Inference on population history and model checking using DNA sequence and micro-satellite data with the software DIYABC (v10). BMC Bioinformatics 11:401
Diekmann OE, Serrao EA (2012) Range-edge genetic diversity: locally poor extant southern patches maintain a regionally diverse hotspot in the seagrass Zostera marina. Mol Ecol 21:1647–1657
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Earl DA, von Holdt BM (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Eckert CG, Samis KE, Lougheed SC (2008) Genetic variation across species’ geographical ranges: the central-marginal hypothesis and beyond. Mol Ecol 17:1170–1188
Eriksson O, Kainulainen K (2011) The evolutionary ecology of dust seeds. Perspect Plant Ecol Evol Syst 33:73–87
Excoffier L, Laval G, Schneider S (2005) ARLEQUIN (version 3.0): An integrated software package for population genetic data analysis. Evol Bioinform Online 1:47–50
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW (2012) Harnessing genomics for delineating conservation units. Trends Ecol Evol 27:489–496
Goldstein DB, Linares AR, Cavalli-Sforza LL, Feldman MW (1995) An evaluation of genetic distances for use with microsatellite loci. Genetics 139:463–471
Goudet J (1995) FSTAT (version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Hammer Ø, Harper DAT, Ryan PD (2001) Past: Paleontological Statistics Software package for education and data analysis. Palaeontol Electron 4:4
Hampe A, Petit RJ (2005) Conserving biodiversity under climate change: the rear edge matters. Ecol Lett 8:461–467
Hampe A, Jump AS (2011) Climate relicts: past, present, future. Ann Rev Ecol Evol Syst 42:313–333
Hewiit GM (2004) Genetic consequences of climatic oscillations in the Quaternary. Phil Trans R Soc Lond B 359:183–195
Hirao AS, Watanabe M, Tsuyuzaki S, Shimono A, Li X, Masuzawa T, Wada N (2017) Genetic diversity within populations of an arctic–alpine species declines with decreasing latitude across the Northern Hemisphere. J Biogeogr 44:2740–2751
Hitchcock CL, Cronquist A (1973) Flora of the Pacific Northwest. University of Washington Press, Seattle
Hynson NA, Mambelli S, Amend AS, Dawson TE (2012) Measuring carbon gains fungal networks in understory plants from the tribe Pyroleae (Ericaceae): a field manipulation and stable isotope approach. Oecologia 169:303–317
Iwasaki T, Aoki K, Seo A, Murakami N (2012) Comparative phylogeography of four component species of deciduous broad-leaved forests in Japan based on chloroplast DNA variation. J Plant Research 125:207–221
Japanese Ministry of the Environment (2015) Red Data Book 2014 - threatened wildlife of Japan, Vol. 8: Vascular plants. GYOSEI Cooperation, Tokyo (in Japan)
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Johansson V, Mikusinska A, Ekblad A, Eriksson O (2015) Partial mycoheterotrophy in Pyloreae: nitrogen and carbon stable isotope signatures during development from seedling to adult. Oecologia 177:203–211
Janes JK, Miller JM, Dupuis JR, Malenfant RM, Gorrell JC, Cullingham CI, Andrew RL (2017) The K=2 conundrum. Mol Ecol 26:3594–3602
Kennedy JP, Preziosi RF, Rowntree JK, Feller IC (2020) Is the central-marginal hypothesis a general rule? Evidence from three distributions of an expanding mangrove species, Avicennia germinans (L.) L. Mol Ecol 29:704–719
Kikuchi A, Miyazaki T, Maki M (2018) Development of microsatellite markers for the endangered semi-shrub Chimaphila umbellata (Ericaceae). Plant Species Biol 33:140–143
Kikuchi R, Pak J-H, Takahashi H, Maki M (2010) Disjunct distribution of chloroplast DNA haplotypes in the understory perennial Veratrum album ssp. oxyseplalum (Melanthiaceae) in Japan due to ancient introgression. New Phytol 187:879–891
Kitamura K, Uchiyama K, Ueno S, Ishizuka W, Tsuyama I, Goto S (2020) Geographical gradients of genetic diversity and differentiation among the southernmost marginal populations of Abies sachalinensis revealed by EST-SSR polymorphism. Forests 11:233
Kortnas J (1972) Corresponding taxa and their ecological background in the forest of temperate Eurasia and North America. In: Valentine DH (ed) Taxonomy, Phytogeography and Evolution. Academic Press, London, pp 37–59
Langella O (1999) Populations 1.2.30: Population genetic software (individuals or population distances, phylogenetic trees). http://bioinformatics.org/~tryphon/populations/
Lascoux M, Palmé A, Cheddadi R, Latta RG (2004) Impact of Ice Ages on the genetic structure of trees and shrubs. Trans R Soc Lond B 359:197–207
Lenoir J, Gégout JC, Marquet PA, De Ruffray P, Brisse H (2008) A significant upward shift in plant species optimum elevation during the 20th century. Science 320:1768–1771
Lesica P, Allendorf FW (1995) When are peripheral populations valuable for conservation? Conserv Biol 9:753–760
Liu L, Wang Z, Huang L, Wang T, Su V (2019) Chloroplast population genetics reveals low levels of genetic variation and conformation to the central-marginal hypothesis in Taxus wallichiana var. mairei, an endangered conifer endemic to China. Ecol Evol 9:11944–11956
Loveless MD, Hamrick JL (1984) Ecological determinants of genetic structure in plant populations. Annl Rev Ecol Syst 15:65–95
Lundell A, Cousins SA, Eriksson O (2015) Population size and reproduction in the declining endangered forest plant Chimaphila umbellata in Sweden. Folia Geobot 50:13–23
Mather LJ, Williams PA (1990) Phenology, seed ecology, and age structure of Spanish health (Erica lusitanica) in Canterbury, New Zealand. N Z J Bot 28:207–216
Nei M, Tajima F, Tateno Y (1983) Accuracy of estimated phylogenetic trees from molecular-data. II. Gene frequency data J Mol Evol 19:153–170
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Newton AC, Allnutt TR, Gillies ACM, Lowe AJ, Ennos RA (1999) Molecular phylogeography, intraspecific variation and the conservation of tree species. Trends Ecol Evol 14:140–145
Parmesan C, Ryrholm N, Stefanescu C, Hill JK, Thomas CD, Descimon H, Huntley B, Kaila L, Kullberg J, Tammaru T, Tennent WJ, Thomas JA, Warren M (1999) Poleward shifts in geographical ranges of butterfly species associated with regional warming. Nature 399:579–578
Parmesan C (2006) Ecological and evolutionary response to recent climate change. Ann Rev Ecol Evol Syst 37:637–669
Peakall ROD, Smouse PE (2006) GENELAX 6: genetic analysis in excel: population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Pérez-Figueroa A, Saura M, Fernández J, Toro MA, Caballero A (2009) METAPOP–a software for the management and analysis of subdivided populations in conservation programs. Conserv Genet 10:1097–1099
Petit RJ, El Mousadik A, Pons O (1998) Identifying populations for conservation on the basis of genetic markers. Conerv Biol 12:844–855
Petit RJ, Aguinagalde I, Beaulieu JL, Bittkau C, Brewer S, Cheddadi R, Ennos R, Fineschi S, Grivet D, Lascoux M, Mohanty A, Müller-Starck D-M, Palmé A, Martín JP, Rendell S, Vendramin GG (2003) Glacial refugia: hotspots but not melting pots of genetic diversity. Science 300:1563–1565
Pironon S, Villellas J, Morris WF, Doak DF, García MB (2015) Do geographic, climatic or historical ranges differentiate the performance of central versus peripheral populations? Global Ecol Biogeogr 24:611–620
Pironon S, Papuga G, Villellas J, Angert AL, García MB, Thompson JD (2016) Geographic variation in genetic and demographic performance: new insights from an old biogeographic paradigm. Biol Rev 92:1877–1909
Piry S, Liukurt G, Cornuet IM (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Hered 90:502–503
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure suing multilocus genotype data. Genetics 155:945–959
Provan J, Bennett KD (2008) Phylogeographic insights into cryptic glacial refugia. Trends Ecol Evol 23:564–571
Qiang D, Fu JZ (2011) When central populations exhibit more genetic diversity than peripheral populations: a simulation study. Chinese Sci Bull 56:2531–2540
Rosenberg NA (2004) Distruct: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) Genepop’007: a computer reimplementation of the Genepop software for Windows and Linux. Mol Ecol Resour 8:103–106
R Development Core Team (2017) R: a language and environment for statistical computing. Version 3.4.0. R Foundation for Statistical Computing. http://www.r-project.org/
Stubbs R, Soltis DE, Cellinese N (2018) The future of cold-adapted plants in changing climates: Micranthes (Saxifragaceae) as a case study. Ecol Evol 8:7164–7177
Sugahara K, Kaneko Y, Sakaguchi S, Ito S, Yamanaka K, Sakio H, Hoshizaki K, Suzuki W, Yamanaka N, Isagi Y, Momohara A, Setoguchi H (2017) Quaternary range-shift history of Japanese wingnut (Pterocarya rhoifokia) in the Japanese Archipelago evidenced from chloroplast DNA and ecological niche modeling. J For Res 22:282–293
Takahashi H (1987) On the infraspecific variations of Chimaphila umbellata (L.) W. Barton (Pyrolaceae). Acta Phytotax Geobot 38:82–96 (in Japanese with an English summary)
Takezaki N, Nei M, Tamura K (2010) POPTREE2: software for constructing population trees from allele frequency data and computing other population statistics with windows interface. Mol Biol Evol 27:747–752
Thuiller W, Lavorel S, Araújo MB, Sykes MT, Prentice IC (2005) Climate change threats to plant diversity in Europe. Proc Natl Acad Sci 102:8245–8250
Thomas CD, Cameron A, Gree RE, Bakkenes M, Beaumont LJ, Collingham YC, Erasmus BFN, de Siqueria MF, Grainger A, Hannah L, Hughes L, Huntley B, van Jaarsveld AS, Midgley GF, Miles L, Ortega-Huerta MA, Peterson AT, Phillips OL, Williams SE (2004) Extinction risk from climate change. Nature 427:145–148
Vucetich JA, Waite TA (2003) Spatial patterns of demography and genetic processes across the species’ range: null hypotheses for landscape conservation genetics. Conserv Genet 6:639–645
Wagner V, Durka W, Hensen I (2011) Increased genetic differentiation but no reduced genetic diversity in peripheral vs. central populations of a steppe grass. Am J Bot 98:1173–1179
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370
Yakimowski X, Eckert CG (2008) Populations do not become less genetically diverse or more differentiated towards the northern limit of the geographical range in clonal Vaccinium stamineum (Ericaceae). New Phytol 180:534–544
Yang H, Li X, Liu D, Chen D, Chen X, Li F, Qi X, Luo Z, Wang C (2016) Genetic diversity and population structure of the endangered medicinal plant Phellodendron amurense in China revealed by SSR marks. Biochem Syst Ecol 66:286–292
Zeng YF, Wang WT, Liao WJ, Wang HF, Zhang DY (2015) Multiple glacial refugia for cool-temperature deciduous trees in northern East Asia: the Mongolian oak as a case study. Mol Ecol 24:5676–5691
Acknowledgements
We would like to thank Drs. S. Horie, Y. Shirosaka, K. Onimaru, A. Uchida, M. Suzuki, J. Yokoyama, and Y. Sakamoto; Messrs. T. Nojima, T. Miyazaki, T. Kimura, T. Kobayashi, and H. Igarashi and Ms. M. Ogishima for material collection. We are also grateful to the regional offices of the Forestry Agency, the Atomic Energy Agency, and Hitachi Seaside Park for permitting sampling. This study was partly supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS).
Funding
This work was partly supported by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science (JSPS) to MM.
Author information
Authors and Affiliations
Contributions
AK and MM collected samples, AK performed molecular experiment, AK and RK performed statistical analyzes, AK and MM conceived the study and wrote the manuscript. All authors approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
There is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Kikuchi, A., Kyan, R. & Maki, M. Population genetic diversity and conservation priority of prince’s pine Chimaphila umbellata populations around the south margin of their distribution. Conserv Genet 22, 839–853 (2021). https://doi.org/10.1007/s10592-021-01366-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01366-x