Abstract
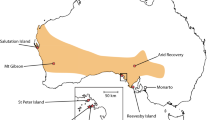
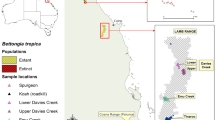
Reintroduction programs can benefit from optimisation of source populations to maximise genetic diversity. Here, we report an approach to guide genetic supplementation of founder individuals to maximise genetic diversity in a reintroduction program for a nationally threatened Australian ground-dwelling marsupial, the southern brown bandicoot (eastern subspecies), Isoodon obesulus obesulus. Following local extinction ~ 100 years earlier, founding individuals were reintroduced to Booderee National Park in south-eastern Australia over three years from the nearest viable wild population, approximately 250 km to the south. To assess and manage risks associated with low genetic diversity, we measured genetic diversity of the reintroduced population relative to the diversity across the subspecies’ range. The genetic diversity of the initial founder population was comparable with other sampled populations; most likely because the source population had high genetic diversity relative to other locations. We simulated scenarios of supplementation of the founder population from alternative candidate sources. We identified populations in the Melbourne region of Victoria as those that would yield the greatest increase in genetic diversity in the Booderee population. The simulated increase in diversity resulting from supplementation was accurately predicted by the pairwise FST comparison between the candidate and recipient population, but not diversity of the supplementation source itself. Genetic diversity is an important consideration in reintroductions; our study enables managers to make informed decisions to maximise the long-term persistence and genetic diversity of reintroduced populations.




Similar content being viewed by others
Data availability
The data generated during our study are available in Dryad, with the following citation and link. Robinson NM, Banks SC (2020) Raw SNP data for Isoodon obesulus obesulus. Dryad. https://doi.org/10.5061/dryad.k98sf7m3g
References
Allendorf FW, Luikart GH, Aitken SN (2012) Conservation and the Genetics of Populations. John Wiley & Sons, Somerset, United Kingdom
Armstrong DP, Seddon PJ (2008) Directions in reintroduction biology. Trends Ecol Evol 23:20–25. https://doi.org/10.1016/j.tree.2007.10.003
Ashby EA, Lunney D, Robertshaw J, Harden R (1990) Distribution and status of bandicoots in New South Wales. In: Brown PR, Wallis RL, Kemper CM (eds) Seebeck JH. Bandicoots and Bilbies. Surrey Beatty & Sons, Sydney
Ballou JD, Lacy RC (1995) Identifying genetically important individuals for management of genetic variation in pedigreed populations. In: Ballou JD, Gilpin M, Foose T (eds) Population management for survival and recovery: analytical methods and strategies in small population conservation. Columbia University Press, New York, pp 76–111
Banes GL, Galdikas BMF, Vigilant L (2016) Reintroduction of confiscated and displaced mammals risks outbreeding and introgression in natural populations, as evidenced by orang-utans of divergent subspecies. Sci Rep 6:22026. https://doi.org/10.1038/srep22026
Cook CN, Sgrò CM (2017) Aligning science and policy to achieve evolutionarily enlightened conservation. Conserv Biol 31:501–512. https://doi.org/10.1111/cobi.12863
Cook CN, Sgrò CM (2018) Understanding managers’ and scientists’ perspectives on opportunities to achieve more evolutionarily enlightened management in conservation. Evol Appl 11:1371–1388. https://doi.org/10.1111/eva.12631
Cook CN, Sgrò CM (2019) Poor understanding of evolutionary theory is a barrier to effective conservation management. Conserv Lett 12:e12619. https://doi.org/10.1111/conl.12619
Cooper S et al (2020) Phylogeography of southern brown and golden bandicoots: implications for the taxonomy and distribution of endangered subspecies and species. Aust J Zool 66:379–393. https://doi.org/10.1071/ZO19052
Cruz VMV, Kilian A, Dierig DA (2013) Development of DArT marker platforms and genetic diversity assessment of the U.S. collection of the new oilseed crop lesquerella and related species. PLoS ONE 8:e64062–e64062. https://doi.org/10.1371/journal.pone.0064062
Dexter N, Macgregor C, Ferguson R (2016) Plan for the translocation of Southern Brown Bandicoots Isoodon obesulus from Nadgee, Timbillica and East Boyd State Forests, New South Wales, to Booderee National Park. Jervis Bay Territory, Department of the Environment
DNP (2015) Booderee National Park Management Plan 2015–2025. Director of National Parks, Canberra
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Fischer MC et al (2017) Estimating genomic diversity and population differentiation–an empirical comparison of microsatellite and SNP variation in Arabidopsis halleri. BMC Genomics. https://doi.org/10.1186/s12864-016-3459-7
Frankham R (1995) Conservation Genetics. Annual Rev Genetics 29:305–327. https://doi.org/10.1146/annurev.ge.29.120195.001513
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618. https://doi.org/10.1111/mec.13139
Frankham R et al (2017) Genetic management of fragmented animal and plant populations. Oxford University Press, Oxford
Frankham R, Briscoe DA, Ballou JD (2002) Introduction to conservation genetics. Cambridge University Press, Cambridge
Frankham R, Ballou JD, Eldridge M, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475. https://doi.org/10.1111/j.1523-1739.2011.01662.x
Frankham R, Bradshaw CJA, Brook BW (2014) Genetics in conservation management: Revised recommendations for the 50/500 rules Red List criteria and population viability analyses. Biol Conserv 170:56–63. https://doi.org/10.1016/j.biocon.2013.12.036
Goudet J (2005) Hierfstat, a package for R to compute and test hierarchical F-statistics. Mol Ecol Notes 5:184–186
Grueber CE, Fox S, McLennan EA, Gooley RM, Pemberton D, Hogg CJ, Belov K (2019) Complex problems need detailed solutions: Harnessing multiple data types to inform genetic management in the wild. Evol Appl 12:280–291. https://doi.org/10.1111/eva.12715
Hasselgren M et al (2018) Genetic rescue in an inbred Arctic fox (Vulpes lagopus) population. Proc R Soc B. https://doi.org/10.1098/rspb.2017.2814
Heber S, Varsani A, Kuhn S, Girg A, Kempenaers B, Briskie J (2013) The genetic rescue of two bottlenecked South Island robin populations using translocations of inbred donors. Proc R Soc B 280:20122228
Hood GM (2010) PopTools version 3.2.5. Available at: http://www.poptools.org [accessed 02/05/2019].
Huff DD, Miller LM, Chizinski CJ, Vondracek B (2011) Mixed-source reintroductions lead to outbreeding depression in second-generation descendents of a native North American fish. Mol Ecol 20:4246–4258. https://doi.org/10.1111/j.1365-294X.2011.05271.x
IUCN / SSC (2013) Guidelines for reintroductions and other conservation translocations. Version 1.0. IUCN Species Survival Commission, Gland, Switzerland
Kilian A et al (2012) Diversity Arrays Technology: a generic genome profiling technology on open platforms. In: Pompanon F, Bonin A (eds) Data production and analysis in population genomics: Methods and protocols. Humana Press, Totowa, NJ, pp 67–89
Lee AM, Engen S, Sæther B-E (2011) The influence of persistent individual differences and age at maturity on effective population size. Proc R Soc B 278:3303–3312. https://doi.org/10.1098/rspb.2011.0283
Li Y, Cooper SJB, Lancaster ML, Packer JG, Carthew SM (2016) Comparative population genetic structure of the endangered southern brown bandicoot Isoodon obesulus, in fragmented landscapes of Southern Australia. PLoS ONE 11:e0152850. https://doi.org/10.1371/journal.pone.0152850
Lindenmayer D, MacGregor C, Dexter N, Fortescue M, Beaton E (2014) Booderee National Park: The Jewel of Jervis Bay. CSIRO Publishing, Collingwood, VIC
Lindenmayer DB et al (2016) Temporal trends in mammal responses to fire reveals the complex effects of fire regime attributes. Ecol Appl 26:557–573. https://doi.org/10.1890/15-0575
Lindenmayer DB et al (2018) Conservation conundrums and the challenges of managing unexplained declines of multiple species. Biol Cons 221:279–292. https://doi.org/10.1016/j.biocon.2018.03.007
Lobert B, Lee A (1990) Reproduction and life history of Isoodon obesulus in Victorian heathland. In: Wallis RL, Kemper CM (eds) Seebeck JH, brown PR. Bandicoots and bilbies. Surrey Beatty & Sons, Sydney, pp 311–318
Marshall TC, Spalton JA (2000) Simultaneous inbreeding and outbreeding depression in reintroduced Arabian oryx. Anim Conserv 3:241–248. https://doi.org/10.1111/j.1469-1795.2000.tb00109.x
McLennan EA, Wright BR, Belov K, Hogg CJ, Grueber CE (2019) Too much of a good thing? Finding the most informative genetic data set to answer conservation questions. Mol Ecol Res 19:659–671. https://doi.org/10.1111/1755-0998.12997
Meek MH et al (2015) Fear of failure in conservation: The problem and potential solutions to aid conservation of extremely small populations. Biol Cons 184:209–217. https://doi.org/10.1016/j.biocon.2015.01.025
Menkhorst P, Knight F (2001) Field guide to the mammals of Australia. Oxford University Press, Melbourne
Menkhorst P, Seebeck JH (1990) Distribution and conservation status of bandicoots in Victoria. In: Seebeck JH, Brown PR, Wallis RL, Kemper CM (eds) Bandicoots and Bilbies. Surrey Beatty & Sons, Sydney, pp 73–84
Miller SA, Dykes DD, Polesky HF (1988) A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res 16:1215–1215. https://doi.org/10.1093/nar/16.3.1215
Moritz C (1999) Conservation units and translocations: strategies for conserving evolutionary processes. Hereditas 130:217–228. https://doi.org/10.1111/j.1601-5223.1999.00217.x
Ottewell K, Dunlop J, Thomas N, Morris K, Coates D, Byrne M (2014) Evaluating success of translocations in maintaining genetic diversity in a threatened mammal. Biol Cons 171:209–219. https://doi.org/10.1016/j.biocon.2014.01.012
Pacioni C, Wayne AF, Page M (2019) Guidelines for genetic management in mammal translocation programs. Biol Cons 237:105–113. https://doi.org/10.1016/j.biocon.2019.06.019
Paull D (1995) The distribution of the southern brown bandicoot (Isoodon obesulus obesulus) in South Australia. Wildl Res 22:585–599
Paull DJ, Mills DJ, Claridge AW (2013) Fragmentation of the southern brown bandicoot Isoodon obesulus: unraveling past climate change from vegetation clearing. Int J Ecol 2013:1–11. https://doi.org/10.1155/2013/536524
Peakall R, Smouse PE (2006) Genalex 6: Genetic analysis in Excel Population genetic software for teaching and research. Mol Ecol Notes 6:288–295. https://doi.org/10.1111/j.1471-8286.2005.01155.x
Pentland C (1999) Population dynamics of the southern brown bandicoot (Isoodon Obesulus) on Ellen Brook Reserve. Edtih Cowan University, Australia
Pierson JC et al (2016) Genetic factors in threatened species recovery plans on three continents. Front Ecol Environ 14:433–440. https://doi.org/10.1002/fee.1323
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ralls K et al (2018) Call for a paradigm shift in the genetic management of fragmented populations. Conserv Lett 11:e12412. https://doi.org/10.1111/conl.12412
Robinson NM, Banks SC (2020) Raw SNP data for Isoodon obesulus obesulus. Dryad. https://doi.org/10.5061/dryad.k98sf7m3g
Robinson NM, MacGregor CI, Hradsky BA, Dexter N, Lindenmayer DB (2018) Bandicoots return to Booderee: initial survival, dispersal, home range and habitat preferences of reintroduced southern brown bandicoots (eastern sub species; Isoodon obesulus obesulus). Wildl Res 45:132–142. https://doi.org/10.1071/WR17040
Strahan R (1995) Mammals of Australia. Smithsonian Institute Press, Washington, D.C.
Taylor HR, Dussex N, van Heezik Y (2017) Bridging the conservation genetics gap by identifying barriers to implementation for conservation practitioners. Global Ecol Conserv 10:231–242. https://doi.org/10.1016/j.gecco.2017.04.001
Travouillon KJ, Phillips M (2018) Total evidence analysis of the phylogenetic relationships of bandicoots and bilbies (Marsupialia: Peramelemorphia): reassessment of two species and description of a new species. Zootaxa 4378:224–256. https://doi.org/10.11646/zootaxa.4378.2.3
Valentine LE, Anderson H, Hardy GES, Fleming PA (2013) Foraging activity by the southern brown bandicoot (Isoodon obesulus) as a mechanism for soil turnover. Aust J Zool 60:419–423. https://doi.org/10.1071/ZO13030
Wahlund S (1928) The combination of populations and the appearance of correlation examined from the standpoint of the study of heredity. Hereditas 11:65–106
Waples RS, Luikart G, Faulkner JR, Tallmon DA (2013) Simple life-history traits explain key effective population size ratios across diverse taxa. Proc R Soc B 280:20131339. https://doi.org/10.1098/rspb.2013.1339
Weeks AR et al (2011) Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol Appl 4:709–725. https://doi.org/10.1111/j.1752-4571.2011.00192.x
Weeks A, Moro D, Thavornkanlapachai R, Taylor R, H, E White N, Weiser E, Heinze D, (2015) Conserving and enhancing genetic diversity in translocation programs. In: Armstrong DP, Hayward MW, Moro D, Seddon PJ (eds) Advances In Reintroduction Biology of Australian and New Zealand Fauna. CSIRO Publishing, Melbourne, pp 127–140
Weeks AR et al (2017) Genetic rescue increases fitness and aids rapid recovery of an endangered marsupial population Nature. Communications 8:1071. https://doi.org/10.1038/s41467-017-01182-3
Westerman M, Kear BP, Aplin K, Meredith RW, Emerling C, Springer MS (2012) Phylogenetic relationships of living and recently extinct bandicoots based on nuclear and mitochondrial DNA sequences. Mol Phylogenet Evol 62:97–108. https://doi.org/10.1016/j.ympev.2011.09.009
White LC, Moseby KE, Thomson VA, Donnellan SC, Austin JJ (2018) Long-term genetic consequences of mammal reintroductions into an Australian conservation reserve. Biol Cons 219:1–11. https://doi.org/10.1016/j.biocon.2017.12.038
Whiteley AR, Fitzpatrick SW, Funk WC, Tallmon DA (2015) Genetic rescue to the rescue. Trends Ecol Evol 30:42–49. https://doi.org/10.1016/j.tree.2014.10.009
Zenger KR, Eldridge MDB, Johnston PG (2005) Phylogenetics, population structure and genetic diversity of the endangered southern brown bandicoot (Isoodon obesulus) in south-eastern Australia. Conserv Genet 6:193–204. https://doi.org/10.1007/s10592-004-7828-4
Acknowledgements
This research received funding from the Australian Government’s National Environmental Science Program through the Threatened Species Recovery Hub. Project partners include Nick Dexter of Booderee National Park (Parks Australia), The Wreck Bay Aboriginal Community Council, Rohan Bilney (Forestry Corporation of New South Wales) and Karrie Rose and Jane Hall of the Taronga Conservation Society. Many thanks to those who provided valuable samples and assisted with analysis for this research, this includes Paul Sunnucks (Monash University), Katherine Harrison (La Trobe University), Sarah Maclagan (Deakin University), Terry Coates (Royal Botanic Gardens Victoria), and The Victorian Government Department of Environment, Land, Water and Planning (DELWP) for the Melbourne region and Glenelg; Andy Murray (DELWP) for the East Gippsland region; Steve Cooper (South Australian Museum) and You Li (Northwest Minzu University) for the Mt. Lofty region; and the Australian Museum for historic samples from the Eden and Mount Gambier regions. Chris MacGregor and other members of the Lindenmayer Long Term Ecology team assisted with the translocation and monitoring.
Funding
This research received funding from the Australian Government’s National Environmental Science Program through the Threatened Species Recovery Hub.
Author information
Authors and Affiliations
Contributions
SCB and NMR conceptualised the study; SCB, CR and NMR designed the study and interpreted results; CR and SCB analysed the data; NMR and CR wrote the manuscript; DBL and JP assisted with interpreting results and editing; All authors gave final approval for publication.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflicts of interest to declare that are relevant to the content of this article.
Ethical approval
Research was approved by the Australian National University Animal Experimentation Ethics Committee (Protocol No. A2015/26) and was undertaken according to the Australian Code of Practice for the Care and Use of Animals for Scientific Purposes.
Consent to participate
This is not applicable to our study. Our research did not require human ethics approval, including consent to participate and publish, as the study did not involve human participation.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Robinson, N.M., Rhoades, C., Pierson, J. et al. Prioritising source populations for supplementing genetic diversity of reintroduced southern brown bandicoots Isoodon obesulus obesulus. Conserv Genet 22, 341–353 (2021). https://doi.org/10.1007/s10592-021-01341-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-021-01341-6