Abstract
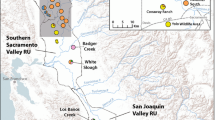
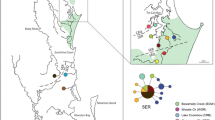
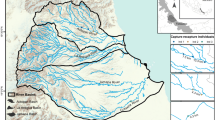
An important consideration when implementing species management is preserving genetic variation, which is fundamental to the long-term persistence of populations and adaptive potential of the species. The European water vole Arvicola amphibius is of high conservation importance in the United Kingdom due to its documented decline in both distribution and abundance. Conservation strategies for this species include protecting source populations, increasing habitat availability and connectivity, non-native predator control and reintroduction. We used mtDNA control region sequences and eight microsatellite markers from samples collected from 12 localities in southeast England, to determine how genetic variation is structured amongst regional water vole populations and to what extent population structure has been influenced by reintroductions. We found high haplotype diversity (h) across native populations in the southeast region and evidence that divergent lineages had been introduced to the region. We detected significant structure between watersheds with mtDNA from native populations and evidence of finer scale structure between populations within watersheds with both mtDNA and microsatellites. We suggest that management strategies should aim to conserve genetic diversity at a population level and that watersheds are a practicable unit for prioritising management within regions. We propose that introductions to restore or augment water voles within watersheds should consider the genetic composition of regional populations and highlight that genetic data has an important role in guiding future conservation options that secure adaptive potential in the face of future environmental change.





Similar content being viewed by others
References
Aars J, Dallas JF, Piertney SB, Marshall F, Gow JL, Telfer S, Lambin X (2006) Widespread gene flow and high genetic variability in populations of water voles Arvicola terrestris in patchy habitats. Mol Ecol 15(6):1455–1466
Allendorf FW, Luikart G (2007) Conservation and the genetics of populations. Blackwell Publishing, Malden
Baker R (2013) Demographic and genetic patterns of water voles in human modified landscapes: implications for conservation. PhD Thesis, University of Brighton, UK
Banes GL, Galdikas BMF, Vigilant L (2016) Reintroduction of confiscated and displaced mammals risks outbreeding and introgression in natural populations, as evidenced by organ-utans of divergent subspecies. Sci Rep 6:22–26
Barreto GR, Rushton SP, Strachan R, Macdonald DW (1998) The role of habitat and mink predation in determining the status of water voles in England. Anim Conserv 2:53–61
Berthier K, Galan M, Foltête JC, Charbonnel N, Cosson JF (2005) Genetic structure of the cyclic fossorial water vole (Arvicola terrestris): Landscape and demographic influences. Mol Ecol 14(9):2861–2871
Biebach I, Keller LF (2012) Genetic variation depends more on admixture than number of founders in reintroduced Alpine ibex populations. Biol Conserv 147(1):197–203
Brace S, Ruddy M, Miller R, Schreve DC, Stewart JR, Barnes I (2016) The colonization history of British water vole (Arvicola amphibius (Linneus, 1758)): Origins and development of the Celtic Fringe. Proc R Soc B 283:20160130
Costello AB, Down TE, Pollard SM, Pacas CJ, Taylor EB (2003) The influence of history and contemporary stream hydrology on the evolution of genetic diversity within species: an examination of microsatellite DNA variation in bull trout, Salvelinus confluentus (Pisces: Salmonidae). Evolution 57(2):328–345
Clement M, Posada D, Crandall KA (2000) TCS: A computer program to estimate gene genealogies. Mol Ecol 9(10):1657–1659
Crandall KA, Templeton AR (1993) Empirical tests of some predictions from coalescent theory with applications to intraspecific phylogeny reconstruction. Genetics 134(3):959–969
Dean M, Strachan R, Gow D, Andrews R (2016) The water vole mitigation handbook. In: Mathews F, Chanin P (eds) The mammal society guidance series. The Mammal Society, London
Dray S, Dufour A (2007) The ade4 package: implementing the duality diagram for ecologists. J Stat Softw 22(4):1–20
Earl DA, vonHoldt BM (2011) Structure Harvester: a website and program for visualizing structure output and implementing the Evanno method. Conserv Genet Resour 4(2):359–361
Edmands S (2007) Between and rock and a hard place: evaluating the relative risks of inbreeding and outbreeding for conservation and management. Mol Ecol 16(3):463–475
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: A simulation study. Mol Ecol 14(8):2611–2620
Excoffier L, Lischer HEL (2010) ARLEQUIN Suite Ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10(3):564–567
Ferguson A (1989) Genetic differences among brown trout, Salmo trutta, stocks and their importance for the conservation and management of the species. Freshw Biol 21:35–46
Frankham R (2005) Genetics and extinction. Biol Conserv 126:131–140
Frankham R (2015) Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol 24:2610–2618
Frankham R, Ballou JD, Eldridge MD, Lacy RC, Ralls K, Dudash MR, Fenster CB (2011) Predicting the probability of outbreeding depression. Conserv Biol 25:465–475
Fisher DO, Lambin X, Yletyinen SM (2008) Experimental translocation of juvenile water voles in a Scottish lowland metapopulation. Popul Ecol 51(2):289–295
Garcia K, Melero Y, Palazón GJ, Castresana J (2017) Spatial mixing of mitochondrial lineages and greater genetic diversity in some invasive populations of the American mink (Neovison vison) compared to native popualtions. Biol Invasions 19(9):2663–2673
Gariboldi MC, Túnez JI, Failla M, Hevia M, Panebianco MV, Viola MNP, Vitullo AD, Cappozzo HL (2016) Patterns of population structure at microsatellite and mitochondrial DNA markers in the franciscana dolphin (Pontoporia blainvillei). Ecol Evol 6:8764–8776
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86(6):485–486
Goudet J (2002) FSTAT, A program to estimate and test gene diversities and fixation indices (Version 2.9.3.2). https://www.unil.ch/izea/softwares/fstat
Hancock JM (1999) Microsatellites and other simple sequences: genomic context and mutational mechanisms. In: Goldstein DB, Schlotterer C (eds) Microsatellites: evolution and applications. Oxford University Press, New York
Holm S (1979) A simple sequentially rejective multiple test procedure. Scand J Stat 6(2):65–70
Houde ALS, Fraser DJ, O’Reilly P, Hutchings JA (2011) Relative risks of inbreeding and outbreeding depression in the wild in endangered salmon. Evol Appl 4(5):634–647
Huff DD, Miller LM, Chizinski CJ, Vondracek B (2011) Mixed-source reintroductions lead to outbreeding depression in second-generation descendants of a native North American fish. Mol Ecol 20:4246–4258
Igea J, Aymerich P, Fernándex-González A, González-Esteban J, Gómez A, Alonso R, Castresana J (2013) Phylogeography and postglacial expansion of the endangered semi-aquatic mammal Galemys pyrenaicus. BMC Evol Biol 13:115
IUCN/SSC (2013) Guidelines for reintroductions and other conservation translocations. Version 1.0. IUCN Species Survival Commission, Gland, Switzerland viiii + 57pp
Jefferies DJ, Morris PA, Mulleneux JE (1989) An enquiry into the changing status of the water vole Arvicola terrestris in Britain. Mamm Rev 19:111–131
JNCC (1995) Biodiversity: the UK Steering Group Report. Volume 2: action plans. HMSO, London
Johnson N (2008) Hybrid incompatibility and speciation. Nat Educ 1(1):20
Johnson JA, Toepfer JE, Dunn PO (2003) Contrasting patterns of mitochondrial and microsatellite population structure in fragmented populations of greater prairie-chickens. Mol Ecol 12:3335–3347
Jombart T (2008) adegenet: an R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jombart T, Devillard S, Dufour AB, Pontier D (2008) Revealing cryptic spatial patterns in genetic variability by a new multivariate method. Heredity 101:92–103
Jost L (2008) G(ST) and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kopelman NM, Mayzel J, Jakobsson M, Rosenberg NA, Mayrose I (2015) CLUMPAK: A program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15(5):1179–1191
Larsson LC, Charlier J, Laikre L, Ryman N (2009) Statistical power for detecting genetic divergence–organelle versus nuclear markers. Conserv Genet 10:1255–1264
Lawson Handley LJ, Perrin N (2007) Advances in our understanding of mammalian sex-biased dispersal. Mol Ecol 16:1559–1578
Lind AJ, Spinks PQ, Fellers GM, Bradley Shaffer H (2011) Rangewide phylogeography and landscape genetics of the Western U.S. endemic frog Rana boylii (Ranidae): implications for the conservation of frogs and rivers. Conserv Genet 12:269–284
Lowe WH, Allendorf FW (2010) What can genetics tell us about population connectivity? Mol Ecol 19:3038–3051
Macdonald DW, Strachan R (1999) The mink and the water vole: analyses for conservation. Wildlife and Conservation Research Unit, Oxford
Manel S, Schwartz MK, Luikart G, Taberlet P (2003) Landscape genetics: combining landscape ecology and population genetics. Trends Ecol Evol 18(4):189–197
McGuire C, Whitfield D (2017) National Water Vole Database and Mapping Project: Part 1 Project Report 2006–2015. Hampshire and Isle of Wight Wildlife Trust, Curdridge
McGuire C, Whitfield D, Perkins H, Owen C (2014) National Water Vole Database and Mapping Project: Guide to the Use of Project Outputs to End of 2012. https://www.hiwwt.org.uk/sites/default/files/files/Reports/Guide%20to%20the%20Use%20of%20Project%20Outputs%202014%20comp.pdf Accessed 4 Feb 2014
Montano V, Jombart T (2017) An Eigenvalue test for spatial principal component analysis. BMC Bioinform 18:562
Moorhouse TP, Gelling M, Macdonald DW (2009) Effects of habitat quality upon reintroduction success in water voles: Evidence from a replicated experiment. Biol Conserv 142:53–60
Moritz C (1994) Defining ‘Evolutionarily Significant Units’ for conservation. Trends Ecol Evol 9(10):373–375
Moritz C (2002) Strategies to protect biological diversity and the evolutionary processes that sustain it. Syst Biol 51(2):238–254
Nei M (1987) Molecular evolutionary genetics. Columbia University Press, New York
Overall ADJ (2016) ConStruct 1.0: an r script to distinguish between substructure and consanguinity within a population using multilocus microsatellite data. Cogent Biol 2(1):1128317
Pasbøll PJ, Berube M, Allendorf FW (2007) Identification of management units using population genetic data. Trends Ecol Evol 22:11–16
Peakall R, Smouse PE (2006) GENALEX 6: Genetic analysis in excel. Population genetic software for teaching and research. Mol Ecol Notes 6(1):288–295
Peakall R, Smouse PE (2012) GENALEX 6.5: genetic analysis in excel. Population genetic software for teaching and research. Bioinformatics 28(19):2537–2539
Pfeiffer I, Völkel I, Täubert H, Brenig B (2004) Forensic DNA-typing of dog hair: DNA-extraction and PCR amplification. Forensic Sci Int 141(2):149–151
Piertney SB, Stewart WA, Lambin X, Telfer S, Aars ON, Dallas JF (2005) Phylogeographic structure and postglacial evolutionary history of water voles (Arvicola terrestris) in the United Kingdom. Mol Ecol 14(5):1435–1444
Pisa G, Orioli V, Spilotros G, Fabbri E, Randi E, Bani L (2015) Detecting a hierarchical genetic population structure: The case study of the fire salamander (Salamandra salamandra) in Northern Italy. Ecol Evol 5(3):743–758
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155(2):945–995
Querejeta M, Fernández-González A, Romero R, Castresana J (2017) Postglacial dispersal patterns and mitochondrial genetic structure of the Pyrenean desman (Galemys pyrenaicus) in the northwestern region of the Iberian Peninsula. Ecol Evol 7:4486–4495
Rambaut A (2006–2014) Figtree: tree figure drawing tool v1.4.0. Institute of Evolutionary Biology Web. https://tree.bio.ed.ac.uk/software/figtree. Accessed 9 July 2014.
Reed DH, Frankham R (2003) Correlation between fitness and genetic diversity. Conserv Biol 17:230–237
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43(1):223–225
Ronquist F, Teslenko M, Van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP (2012) MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61(3):539–542
Rosenberg NA, Burke T, Elo K et al (2001) Empirical evaluation of genetic clustering methods using multilocus genotypes from 20 chicken breeds. Genetics 159(2):699–713
Ryder OA (1986) Species conservation and systematics: the dilema of subspecies. Trends Ecol Evol 1:9–10
Schmitt T (2007) Molecular biogeography of Europe: Pleistocene cycles and postglacial trends. Front Zool 4:11
Schwartz MK, Luikart G, Waples RS (2005) Genetic monitoring as a promising tool for conservation and management. Trends Ecol Evol 22:25–33
Senn H, Ogden R, Frosch C et al (2014) Nuclear and mitochondrial genetic structure in the Eurasian beaver (Castor fiber)—implications for future reintroductions. Evol Appl 7:645–662
Simpson S, Blampied N, Peniche G, Dozières A, Blankett T, Coleman S, Cornish N, Groombridge JJ (2013) Genetic structures of introduced populations: 120-year-old DNA footprint of historic introduction in an insular small mammal population. Ecol Evol 3(3):614–628
Stewart WA, Piertney SB, Dallas JF (1998) Isolation and characterization of highly polymorphic microsatellites in the water vole, Arvicola terrestris. Mol Ecol 7:1258–1259
Stoddart M (1970) Individual range, dispersion and dispersal in a population of water voles (Arvicola terrestris (L.). J Anim Ecol 37:403–424
Strachan R, Jefferies DJ (1993) The water vole Arvicola terrestris in Britain 1989–1990: its distribution and changing status. The Vincent Wildlife Trust, London
Strachan C, Strachan R, Jefferies DJ (2000) Preliminary report on the changes in the water vole as shown by the National Surveys of 1989–1990 and 1996–1998. The Vincent Wildlife Trust, London
Strachan R, Moorhouse T, Gelling M (2011) Water vole conservation handbook, 3rd edn. The Wildlife Conservation Unit, Abingdon
Stewart WA, Dallas JF, Piertney SB, Marshall F, Lambin X, Telfer S (1999) Metapopulation genetic structure in the water vole, Arvicola terrestris, in NE Scotland. Biol J Lin Soc 68:159–171
Stucky BJ (2012) Seqtrace: A graphical tool for rapidly processing DNA sequencing chromatograms. J Biomol Tech 23(3):90–93
Tajima F (1989) Statistical method for testing the neutral mutation hypothesis by DNA polymorphism. Genetics 123:585–595
Telfer S, Piertney SB, Dallas JF, Stewart WA, Marshal F, Gow J, Lambin X (2003) Parentage assignment reveals widespread and large-scale dispersal in water voles. Mol Ecol 12:1939–1951
Van-Oosterhout C, Hutchinson WF, Wills DPD, Shipley P (2004) Micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4(3):535–538
Vergara M, Basto MP, Madeira MJ, Gómez-Moliner BJ, Santos-Reis M, Fernandes C, Ruiz-González A (2015) Inferring population genetic structure in widely and continuously distributed carnivores: the stone marten as a case study. PLoS ONE 10(7):e0134257
Verkuil YI, Juillet C, Lank DB, Widemo F, Piersma T (2014) Genetic variation in nuclear and mitochondrial markers supports a large sex difference in lifetime reproductive skew in a lekking species. Ecol Evol 4(18):3626–3632
Wan QH, Wu H, Fujihara T, Fang SG (2004) Which genetic marker for which conservation genetics issue? Electrophoresis 25:2165–2176
Waples RS, Gagiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15(6):1419–1439
Weeks AR, Sgro CM, Young AG, Frankham R, Mitchell NJ, Miller KA, Byrne M, Coates DJ, Eldridge MDB, Sunnucks P, Breed MR, James EA, Hoffmann AA (2011) Assessing the benefits and risks of translocations in changing environments: a genetic perspective. Evol Appl 4(6):709–725
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38(6):1358–1370
Woodroffe GL, Lawton JH, Davidson WL (1990) The impact of American mink Mustela vison on water voles Arvicola terrestris in the North Yorkshire Moors National Park. Biol Conserv 51:49–62
Acknowledgements
We are grateful to all the landowners and volunteers for support with field work. In particular, we would like to thank Chloe Sadler and Fran Southgate from the Wildlife Trusts, Paul Stevens and Adam Salmon from the Wildfowl and Wetlands Trust, and Jane Reeves and volunteers from the Manhood Wildlife Trust and Heritage Group. We would also like to acknowledge Hampshire Biodiversity Information Centre, Hampshire Mammal Group, Kent Biodiversity Records Centre, Kent Mammal Group and Greenspace Information for Greater London for kindly providing distributional data on water voles. Lastly, we would also like to thank Inga Zeisset from the University of Brighton for her valuable comments on the manuscript. This project was part funded by the Arun & Rother Connections Heritage Lottery Fund, the Environment Agency and the Royal Geographical Society with IBS Postgraduate Research Award.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Baker, R.J., Scott, D.M., King, P.J. et al. Genetic structure of regional water vole populations and footprints of reintroductions: a case study from southeast England. Conserv Genet 21, 531–546 (2020). https://doi.org/10.1007/s10592-020-01268-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-020-01268-4