Abstract
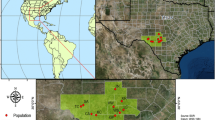
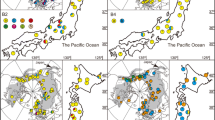
Understanding patterns of speciation and subsequent gene flow can clarify the evolutionary origins and history of species endemic to a specific geographic region and reveal genetic patterns important for conservation and management of rare species. We chose Dicerandra from the North American Coastal Plain biodiversity hotspot as a model to explore these concepts because of its endemism and the threatened status of most of its species. Using microsatellite-based population-level analyses of 32 populations from four of the annual species (D. linearifolia var. linearifolia, D. linearifolia var. robustior, D. fumella, D. odoratissima, and D. radfordiana), we addressed questions of genetic diversity, population structure, and hybridization. Strong support was found for the species-level recognition of the recently described D. fumella from the Florida panhandle. Dicerandra linearifolia var. linearifolia showed some regional cohesion of populations, but there was no consistent geographic pattern to the clustering of populations. Dicerandra radfordiana showed consistent clustering with proximate populations of D. odoratissima. Given that D. radfordiana is found at the southeastern extreme of the range of D. odoratissima, these populations may represent the early stages of speciation by isolation. While there are morphological and bioclimatic niche distinctions between D. odoratissima and D. radfordiana, there is no molecular support for a distinct D. radfordiana. Overall, there is modest genetic diversity found at the population level for all Dicerandra annuals. Microsatellite data support previously proposed hypotheses of hybridization between D. linearifolia var. linearifolia and D. odoratissima, but do not support such hypotheses for D. fumella and D. linearifolia var. robustior.







Similar content being viewed by others
References
Allen JM, Germain-Aubrey CC, Barve N, Neubig KM, Majure LC, Laffan SW, Mishler BD, Owens HL, Smith SA, Whitten WM, Abbott JR, Soltis DE, Guralnick R, Soltis PS (2019) Spatial phylogenetics of Florida vascular plants: the effects of calibration and uncertainty on diversity estimates. iScience 11:57–70
Baldwin BG (2007) Adaptive radiation of shrubby tarweeds (Deinandra) in the California Islands parallels diversification of the Hawaiian silversword alliance (Compositae–Madiinae). Am J Bot 94:237–248
Ball MC, Finnegan L, Manseau M et al (2010) Integrating multiple analytical approaches to spatially delineate and characterize genetic population structure: an application to boreal caribou (Rangifer tarandus caribou) in central Canada. Conserv Genet 11:21–31
Blischak P, Chifman J, Wolfe AD, Kubatko LS (2018) HyDe: a python package for genome-scale hybridization detection. Syst Biol 67:821–829
Castoe T, Poole A, GU W et al (2010) Rapid identification of thousands of copperhead snake (Agkistrodon contortrix) microsatellite loci from modest amounts of 454 shotgun genome sequence. Mol Ecol Resour 10:341–347
Christman SP, Judd WS (1990) Notes on plants endemic to Florida scrub. Fla Sci 53:52–73
Clivati D, Gitzendanner MA, Hilsdorf AWS, Araújo WL, Miranda VFO (2012) Microsatellite markers developed for Utricularia reniformis (Lentibulariaceae). Am J Bot 99:375–378
De Queiroz K (2007) Species concepts and species delimitation. Syst Biol 56:879–886
Degnan JH (2018) Modeling hybridization under the network multispecies coalescent. Syst Biol 67:786–799
Earl D, VonHoldt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the method. Conserv Genet Resour 4:359–361
Edwards C, Soltis D, Soltis P (2008) Using patterns of genetic structure based on microsatellite loci to test hypotheses of current hybridization, ancient hybridization and incomplete lineage sorting in Conradina (Lamiaceae). Mol Ecol 17:5157–5174
Estill J, Cruzan M (2001) Phytogeography of rare plant species endemic to the southeastern United States. Castanea 66:3–23
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer H (2010) Arlequin suite ver 3.5: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Resour 10:564–567
Folk RA, Soltis P, Soltis D, Guralnick R (2018a) New prospects in the detection and comparative analysis of hybridization in the tree of life. Am J Bot 105:364–375
Folk R, Visger C, Soltis P, Soltis D, Guralnick R (2018b) Geographic range dynamics drove ancient hybridization in a lineage of angiosperms. Am Nat 192:171–187
Givnish et al (2008) Origin, adaptive radiation and diversification of the Hawaiian lobeliads (Asterales: Campanulaceae). Proc R Soc B 276:407–416
Gordon A (2008) Fastx-toolkit. http://hannonlab.cshl.edu/fastx_toolkit. Accessed Jan 2019
Hoban SM, Hauffe HC, Pérez-Espona S et al (2013) Bringing genetic diversity to the forefront of conservation policy and management. Conserv Genet Resour 5:593–598
Huck R (1987) Systematics and evolution of Dicerandra (Labiatae). Phanerogamarum Monogr 19:1–343
Huck R (2010) Dicerandra fumella (Lamiaceae), a new species in the Florida panhandle and adjacent Alabama, with comments on the D. linearifolia complex. Rhodora 112:215–227
Jakobsson M, Rosenberg N (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806
Jombart T (2008) Adegenet: a R package for the multivariate analysis of genetic markers. Bioinformatics 24:1403–1405
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Jost L, Archer F, Flanagan S, Gaggiotti O, Hoban S, Latch E (2017) Differentiation measures for conservation genetics. Evol Appl 11:1139–1148
Kalinowski ST (2011) The computer program STRUCTURE does not reliably identify the main genetic clusters within species: simulations and implications for human population structure. Heredity 106:625–632
Kruckeberg A, Rabinowitz D (1985) Biological aspects of endemism in higher plants. Annu Rev Ecol Syst 16:447–479
Laikre L (2010) Genetic diversity is overlooked in international conservation policy implementation. Conserv Genet 11:349–354
Laikre L et al (2010) Neglect of genetic diversity in implementation of the convention of biological diversity. Conserv Biol 24:86–88
Meirmans PG (2015) Seven common mistakes in population genetics and how to avoid them. Mol Ecol 24:3223–3231
Meirmans P, Hedrick P (2011) Assessing population structure: FST and related measures. Mol Ecol Resour 11:5–18
Meirmans P, Van Tiederen P (2004) GENOTYPE and GENODIVE: two programs for the analysis of genetic diversity of asexual organisms. Mol Ecol Notes 4:792–794
Menges E (1992) Habitat preferences and response to disturbance for Dicerandra frutescens, a Lake Wales Ridge (Florida) endemic plant. Bull Torrey Bot Club 119:308–313
Myers R, Ewel J (1990) Ecosystems of Florida. Myers and Ewel (ed). University Presses of Florida, Gainesville, pp 35–69, 150–193
Nosil P (2008) Speciation with gene flow could be common. Mol Ecol 19:2103–2106
Noss RF, Platt WJ, Sorrie BA, Weakley AS, Means DB, Costanza J, Peet RK (2015) How global biodiversity hotspots may go unrecognized: lessons from the North American Coastal Plain. Divers Distrib 21:236–244
Oliveira LO, Huck RB, Gitzendanner MA, Judd WS, Soltis DE, Soltis PS (2007) Molecular phylogeny, biogeography, and systematics of Dicerandra (Lamiaceae), a genus endemic to the southeastern United States. Am J Bot 94:1017–1027
Peakall R, Smouse P (2006) GENALEX 6: genetic analysis in Excel. Population genetic software for teaching and research. Mol Ecol Notes 6:288–295
Petit RJ, Excoffier L (2009) Gene flow and species delimitation. Trends Ecol Evol 24:386–393
Pollock LJ, Rosauer DF, Thornhill AH et al (2015) Phylogenetic diversity meets conservation policy: small areas are key to preserving eucalypt lineages. Philos Trans R Soc B 370:20140007
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rieseberg L, Soltis D (1991) Phylogenetic consequences of cytoplasmic gene flow in plants. Evol Trends Plants 5:65–84
Rosenberg N (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rousset F (2008) GENEPOP: a complete re-implementation of the GENEPOP software for Windows and Linux. Mol Ecol Resour 8:103–106
Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. In: Misener S, Krawetz SA (eds) Bioinformatics methods and protocols. Methods in molecular biology, vol 132. Humana Press, Totowa
Schuelke M (2000) An economic method for the fluorescent labeling of PCR fragments. Nat Biotechnol 18:223–224
Swafford D (2003) PAUP* phylogenetic analysis using parsimony (*and other methods), version 4. Sinauer Associates, Sunderland
USFWS (2007) Endangered and threatened wildlife and plants; 5-year review of 22 southeastern species. Fed Reg 72:20866–20868
Wendel J, Doyle J (1998) Phylogenetic incongruence: window into genome history and molecular evolution. In: Soltis S, Doyle JJ (eds) Molecular systematics of plants II. Kluwer Academic Publishers, Norwell, pp 265–296
Wheeler Q, Meier R (2000) Species concepts and phylogenetic theory. Columbia University Press, West Sussex
Wunderlin RP, Hanson BF (2011) Guide to the vascular plants of Florida, 3rd edn. University Press of Florida, Gainesville
Acknowledgements
This work was supported by research grants from the Florida Native Plant Society and the University of Florida Biology Department (to A.C.P.). Additional support was provided by the National Science Foundation Graduate Research Fellowship under Grant No. DGE-1315138 and DGE-1842473 (to A.A.N).
Author information
Authors and Affiliations
Corresponding authors
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Payton, A.C., Naranjo, A.A., Judd, W. et al. Population genetics, speciation, and hybridization in Dicerandra (Lamiaceae), a North American Coastal Plain endemic, and implications for conservation. Conserv Genet 20, 531–543 (2019). https://doi.org/10.1007/s10592-019-01154-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-019-01154-8