Abstract
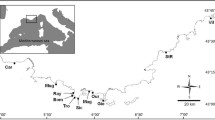
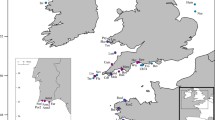
The discovery of two genetically distinct splittail populations within the San Francisco Estuary, one which spawns in the rivers of the Central Valley and another in the Petaluma and Napa Rivers of the San Pablo Bay, prompted the need to evaluate their degree of connectivity and relative sizes. We genotyped multiple age-0 splittail cohorts using 19 microsatellite loci to assess any spatiotemporal changes in the distribution of the two populations and estimate their effective population sizes (N e). Genetic population assignments demonstrated that while age-0 splittail are predominantly spatially segregated by populations, substantial geographical overlap may occur during years of high precipitation. However, despite this periodic range overlap, the original observed population structure has persisted for nearly a decade which has included a similarly wet year. This suggests that the present population structure will likely persist in the future due to strong philopatry and/or adaptive differences. We also found that N e estimates were generally lower for the San Pablo Bay population than the Central Valley population, which is consistent with the relative amount of habitat availability in the two locations and genetic diversity indices. The relative isolation and apparent lower N e of the San Pablo Bay splittail population indicates a higher vulnerability to extinction. A more consistent monitoring effort of splittail in the Petaluma and Napa Rivers may be necessary in order to better understand the future viability of this less studied population.





Similar content being viewed by others
References
Antao T, Perez-Figueroa A, Luikart G (2011) Early detection of population declines: high power of genetic monitoring using effective population size estimators. Evol Appl 4:144–154. doi:10.1111/j.1752-4571.2010.00150.x
Baerwald MR, May B (2004) Characterization of microsatellite loci for five members of the minnow family Cyprinidae found in the Sacramento-San Joaquin Delta and its tributaries. Mol Ecol Notes 4:385–390. doi:10.1111/j.1471-8286.2004.00661.x
Baerwald M, Bien V, Feyrer F, May B (2007) Genetic analysis reveals two distinct Sacramento splittail (Pogonichthys macrolepidotus) populations. Conserv Genet 8:159–167. doi:10.1007/s10592-006-9157-2
Baerwald MR, Feyrer F, May B (2008) Distribution of genetically differentiated splittail populations during the nonspawning season. Trans Am Fish Soc 137:1335–1345. doi:10.1577/T07-097.1
Charlesworth B (2009) Effective population size and patterns of molecular evolution and variation. Nat Rev Genet 10:195–205. doi:10.1038/nrg2526
Cloern JE, Knowles N, Brown LR et al (2011) Projected evolution of California’s San Francisco Bay-Delta-river system in a century of climate change. PLoS ONE 6:e24465. doi:10.1371/journal.pone.0024465
Coombs JA, Letcher BH, Nislow KH (2012) GONe: software for estimating effective population size in species with generational overlap. Mol Ecol Resour 12:160–163. doi:10.1111/j.1755-0998.2011.03057.x
Crawford NG (2010) SMOGD: software for the measurement of genetic diversity. Mol Ecol Resour 10:556–557. doi:10.1111/j.1755-0998.2009.02801.x
Daniels RA, Moyle PB (1983) Life history of splittail (Cyprinidae: Pogonichthys macrolepidotus) in the Sacramento-San Joaquin Estuary. Fish Bull 81:647–654
Duong TY, Scribner KT, Forsythe PS et al (2013) Interannual variation in effective number of breeders and estimation of effective population size in long-lived iteroparous lake sturgeon (Acipenser fulvescens). Mol Ecol 22:1282–1294. doi:10.1111/mec.12167
Earl DA, VonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
England PR, Luikart G, Waples RS (2010) Early detection of population fragmentation using linkage disequilibrium estimation of effective population size. Conserv Genet 11:2425–2430. doi:10.1007/s10592-010-0112-x
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. doi:10.1111/j.1365-294X.2005.02553.x
Excoffier L, Laval G, Schneider S (2005) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evol Bioinform Online 1:47–50
Feyrer F, Sommer TR, Baxter RD (2005) Spatial-temporal distribution and habitat associations of age-0 splittail in the lower San Francisco Estuary watershed. Copeia 1:159–168
Feyrer F, Sommer T, Hobbs J (2007) Living in a dynamic environment: variability in life history traits of age-0 splittail in tributaries of San Francisco Bay. Trans Am Fish Soc 136:1393–1405. doi:10.1577/T06-253.1
Feyrer F, Hobbs J, Sommer T (2010) Salinity inhabited by age-0 splittail (Pogonichthys macrolepidotus) as determined by direct field observation and retrospective analyses with otolith chemistry. San Franc Estuary Watershed Sci 8: Article 2
Franklin IR (1980) Evolutionary change in small populations. In: Soule ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer Associates, Sunderland, pp 135–149
Franklin IR, Frankham R (1998) How large must populations be to retain evolutionary potential? Anim Conserv 1:69–73
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Hill WG (1981) Estimation of effective population size from data on linkage disequilibrium. Genet Res 38:209–216
Hobbs J (2013) Age determination of San Pablo Bay splittail from otoliths and scales. University of California, Davis
Jakobsson M, Rosenberg NA (2007) CLUMPP: a cluster matching and permutation program for dealing with label switching and multimodality in analysis of population structure. Bioinformatics 23:1801–1806. doi:10.1093/bioinformatics/btm233
Jorde PE, Ryman N (1995) Temporal allele frequency change and estimation of effective size in populations with overlapping generations. Genetics 139:1077–1090
Jorde PE, Ryman N (2007) Unbiased estimator for genetic drift and effective population size. Genetics 177:927–935. doi:10.1534/genetics.107.075481
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026. doi:10.1111/j.1365-294X.2008.03887.x
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189. doi:10.1111/j.1471-8286.2004.00845.x
Kimmerer WJ (2002) Effects of freshwater flow on abundance of estuarine organisms: physical effects or trophic linkages? Mar Ecol Prog Ser 243:39–55. doi:10.3354/meps243039
Kimmerer W (2004) Open water processes of the San Francisco Estuary: from physical forcing to biological responses. San Franc Estuary Watershed Sci 2: Article 1
Luikart G, Ryman N, Tallmon DA et al (2010) Estimation of census and effective population sizes: the increasing usefulness of DNA-based approaches. Conserv Genet 11:355–373. doi:10.1007/s10592-010-0050-7
Lynch M, Lande R (1998) The critical effective size for a genetically secure population. Anim Conserv 1:70–72
Mahardja B, May B, Baerwald MR (2012) Characterization of 36 additional microsatellite loci in splittail (Pogonichthys macrolepidotus) and cross-amplification in five other native Californian cyprinid species. Conserv Genet Resour 4:917–921. doi:10.1007/s12686-012-9673-y
Moyle PB (2002) Inland fishes of California. University of California Press, Berkeley
Moyle PB, Baxter RD, Sommer T, et al. (2004) Biology and population dynamics of Sacramento splittail (Pogonichthys macrolepidotus) in the San Francisco Estuary: a review. San Franc Estuary Watershed Sci 2: Article 3
Nei M, Tajima F (1981) Genetic drift and estimation of effective population size. Genetics 98:625–640
Peery MZ, Kirby R, Reid BN et al (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418. doi:10.1111/j.1365-294X.2012.05635.x
Pollak E (1983) A new method for estimating the effective population size from allele frequency changes. Genetics 104:531–548
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Raymond M, Rousset F (1995) GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Rice WR (1989) Analyzing tables of statistical tests. Evolution 43:223–225
Serbezov D, Jorde PE, Bernatchez L et al (2012) Life history and demographic determinants of effective/census size ratios as exemplified by brown trout (Salmo trutta). Evol Appl 5:607–618. doi:10.1111/j.1752-4571.2012.00239.x
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Sommer T, Baxter R, Herbold B (1997) Resilience of splittail in the Sacramento-San Joaquin Estuary. Trans Am Fish Soc 126:961–976
Sommer TR, Baxter RD, Feyrer F (2007) Splittail “delisting”: a review of recent population trends and restoration activities. Am Fish Soc Symp 53:25–38
USFWS (2010) Endangered and threatened wildlife and plants; 12-month finding on a petition to list the Sacramento splittail as endangered or threatened. 62070–62093. United States Fish and Wildlife Service
Vähä J-P, Primmer CR (2006) Efficiency of model-based Bayesian methods for detecting hybrid individuals under different hybridization scenarios and with different numbers of loci. Mol Ecol 15:63–72. doi:10.1111/j.1365-294X.2005.02773.x
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) Micro-checker: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538. doi:10.1111/j.1471-8286.2004.00684.x
Wang J (2001) A pseudo-likelihood method for estimating effective population size from temporally spaced samples. Genet Res 78:243–257. doi:10.1017/S0016672301005286
Wang J (2009) A new method for estimating effective population sizes from a single sample of multilocus genotypes. Mol Ecol 18:2148–2164. doi:10.1111/j.1365-294X.2009.04175.x
Wang J, Whitlock MC (2003) Estimating effective population size and migration rates from genetic samples over space and time. Genetics 163:429–446
Waples RS (1989) A generalized approach for estimating effective population size from temporal changes in allele frequency. Genetics 121:379–391
Waples RS (2005) Genetic estimates of contemporary effective population size: to what time periods do the estimates apply? Mol Ecol 14:3335–3352. doi:10.1111/j.1365-294X.2005.02673.x
Waples RS (2006) A bias correction for estimates of effective population size based on linkage disequilibrium at unlinked gene loci. Conserv Genet 7:167–184. doi:10.1007/s10592-005-9100-y
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Resour 8:753–756. doi:10.1111/j.1755-0998.2007.02061.x
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol Appl 3:244–262. doi:10.1111/j.1752-4571.2009.00104.x
Waples RS, England PR (2011) Estimating contemporary effective population size on the basis of linkage disequilibrium in the face of migration. Genetics 189:633–644. doi:10.1534/genetics.111.132233
Waples RS, Yokota M (2007) Temporal estimates of effective population size in species with overlapping generations. Genetics 175:219–233. doi:10.1534/genetics.106.065300
Waples RS, Luikart G, Faulkner JR, Tallmon DA (2013) Simple life-history traits explain key effective population size ratios across diverse taxa. Proc R Soc B 280:20131339
Willi Y, Van Buskirk J, Hoffmann AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Evol Syst 37:433–458. doi:10.2307/annurev.ecolsys.37.091305.30000017
Wright S (1931) Evolution in Mendelian populations. Genetics 16:97–159
Young PS, Cech JJ (1996) Environmental tolerances and requirements of splittail. Trans Am Fish Soc 125:664–678
Acknowledgments
We would like to thank Kyle Howarth and Katja Waldron for laboratory work support; James Hobbs for sharing his findings regarding splittail age structure in the San Pablo Bay; Matthew Young and Molly Stephens for providing assistance with the preparation of the manuscript. We also thank two anonymous reviewers who provided helpful comments that improved this manuscript.
Funding
This material is based upon work supported by the Delta Science Program under Grant No. 2037.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mahardja, B., May, B., Feyrer, F. et al. Interannual variation in connectivity and comparison of effective population size between two splittail (Pogonichthys macrolepidotus) populations in the San Francisco Estuary. Conserv Genet 16, 385–398 (2015). https://doi.org/10.1007/s10592-014-0665-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0665-1