Abstract
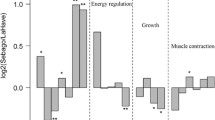
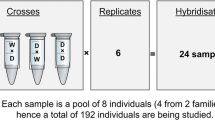
One of the major challenges facing conservation biology is characterizing the genetic variation underlying adaptation to different environments. Gene expression is the process whereby genomic information is converted into phenotype and quantitative variation in gene expression is linked to phenotypic variation. Identifying gene transcription profiles that provide fitness benefits in specific environments would promote more effective species reintroduction and conservation practices. In this study, we developed a custom oligonucleotide microarray for Atlantic salmon (Salmo salar) and used this microarray to measure gene transcription in gill tissue for two Atlantic salmon strains currently being reintroduced into Lake Ontario: LaHave (anadromous) and Sebago (landlocked). We measured gene transcription in juvenile salmon from each strain that had been reared under the same conditions and identified genes differentially expressed between the two strains. We used the normalized transcription data and microsatellite genotype data to partition the variance into effects of selection versus genetic drift. We found that although there was little genetic differentiation (F ST = 0.038) between the two strains, 21 genes were significantly differentially expressed between the two strains, and in all cases the difference was consistent with divergence by selection. We use this analysis to predict the Sebago strain will be more likely to be successfully reintroduced, highlighting how the combination of population genetics with gene expression can help to guide reintroduction efforts.


Similar content being viewed by others
References
Aykanat T, Thrower FP, Heath DD (2011) Rapid evolution of osmoregulatory function by modification of gene transcription in steelhead trout. Genetica 139:233–242. doi:10.1007/s10709-010-9540-2
Baayen RH (2008) Analyzing linguistic data: a practical introduction to statistics using R. Cambridge University Press, Cambridge, UK, New York
Banka CL, Yuan T, de Beer MC et al (1995) Serum amyloid A (SAA): influence on HDL-mediated cellular cholesterol efflux. J Lipid Res 36:1058–1065
Bates D, Maechler M (2009) lme4: Linear mixed-effects models using {S4} classes.{R} package version 0.999375-32
Bobrowski R (2010) Survival, growth, and out-migration timing of reintroduced Atlantic salmon (Salmo salar) in Cobourg Brook, Ontario. M.Sc., Trent University (Canada)
Campos-Perez JJ, Ward M, Grabowski PS et al (2000) The gills are an important site of iNOS expression in rainbow trout Oncorhynchus mykiss after challenge with the gram-positive pathogen Renibacterium salmoninarum. Immunology 99:153–161
Chatzigeorgiou A, Lyberi M, Chatzilymperis G et al (2009) CD40/CD40L signaling and its implication in health and disease. BioFactors Oxf Engl 35:474–483. doi:10.1002/biof.62
Coghlan SM, Ringler NH (2004) Comparison of Atlantic salmon embryo and fry stocking in the salmon river, New York. North Am J Fish Manag 24:1385–1397. doi:10.1577/M03-253.1
Colotta F, Re F, Muzio M et al (1993) Interleukin-1 type II receptor: a decoy target for IL-1 that is regulated by IL-4. Science 261:472–475
Crawford SS (2001) Salmonine introductions to the Laurentian Great Lakes: an historical review and evaluation of ecological effects. NRC Research Press, Ottawa
Debes PV, Normandeau E, Fraser DJ et al (2012) Differences in transcription levels among wild, domesticated, and hybrid Atlantic salmon (Salmo salar) from two environments. Mol Ecol 21:2574–2587. doi:10.1111/j.1365-294X.2012.05567.x
Dimond P, Smitka J (2005) Evaluation of selected strains of Atlantic salmon as potential candidates for the restoration of Lake Ontario. Trout Unlimited Canada Technical Report ON-12. Guelph, Ontario: Trout Unlimited Canada
Essex-Fraser PA, Steele SL, Bernier NJ et al (2005) Expression of four Glutamine Synthetase Genes in the Early Stages of Development of Rainbow Trout (Oncorhynchus mykiss) in Relationship to Nitrogen Excretion. J Biol Chem 280:20268–20273. doi:10.1074/jbc.M412338200
Evans DH, Piermarini PM, Choe KP (2005) The multifunctional fish gill: dominant site of gas exchange, osmoregulation, acid-base regulation, and excretion of nitrogenous waste. Physiol Rev 85:97–177. doi:10.1152/physrev.00050.2003
Frankham R (2003) Genetics and conservation biology. C R Biol 326(Supplement 1):22–29. doi:10.1016/S1631-0691(03)00023-4
Frankham R, Briscoe DA, Ballou JD (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
Gaboriaud C, Thielens NM, Gregory LA et al (2004) Structure and activation of the C1 complex of complement: unraveling the puzzle. Trends Immunol 25:368–373. doi:10.1016/j.it.2004.04.008
Garcia de Leaniz C, Fleming IA, Einum S et al (2007) A critical review of adaptive genetic variation in Atlantic salmon: implications for conservation. Biol Rev 82:173–211. doi:10.1111/j.1469-185X.2006.00004.x
Giger T, Excoffier L, Day PJR et al (2006) Life history shapes gene expression in salmonids. Curr Biol CB 16:R281–282. doi:10.1016/j.cub.2006.03.053
Giger T, Excoffier L, Amstutz U et al (2008) Population transcriptomics of life-history variation in the genus Salmo. Mol Ecol 17:3095–3108. doi:10.1111/j.1365-294X.2008.03820.x
Goldrath AW, Bevan MJ (1999) Selecting and maintaining a diverse T-cell repertoire. Nature 402:6–13. doi:10.1038/35005508
Goltry KL, Epperly MW, Greenberger JS (1998) Induction of serum amyloid A inflammatory response genes in irradiated bone marrow cells. Radiat Res 149:570–578
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Hered 86:485–486
Grishkevich V, Yanai I (2013) The genomic determinants of genotype × environment interactions in gene expression. Trends Genet TIG 29:479–487. doi:10.1016/j.tig.2013.05.006
Hansen MM (2010) Expression of interest: transcriptomics and the designation of conservation units. Mol Ecol 19:1757–1759. doi:10.1111/j.1365-294X.2010.04597.x
Hedrick PW (2001) Conservation genetics: where are we now? Trends Ecol Evol 16:629–636. doi:10.1016/S0169-5347(01)02282-0
Houde AL, Wilson CC, Neff BD (2013) Genetic architecture of survival and fitness-related traits in two populations of Atlantic salmon. Heredity. doi:10.1038/hdy.2013.74
Houde ALS, Wilson CC, Neff BD (2014) Competitive interactions among multiple non-native salmonids and two populations of Atlantic salmon. Ecol Freshw Fish n/a–n/a. doi: 10.1111/eff.12123
Huang DW, Sherman BT, Lempicki RA (2008) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4:44–57. doi:10.1038/nprot.2008.211
Hutter S, Saminadin-Peter SS, Stephan W, Parsch J (2008) Gene expression variation in African and European populations of Drosophila melanogaster. Genome Biol 9:R12. doi:10.1186/gb-2008-9-1-r12
Lamaze FC, Garant D, Bernatchez L (2013) Stocking impacts the expression of candidate genes and physiological condition in introgressed brook charr (Salvelinus fontinalis) populations. Evol Appl 6:393–407. doi:10.1111/eva.12022
Larsen PF, Nielsen EE, Williams TD et al (2007) Adaptive differences in gene expression in European flounder (Platichthys flesus). Mol Ecol 16:4674–4683. doi:10.1111/j.1365-294X.2007.03530.x
Lin B, Chen S, Cao Z et al (2007) Acute phase response in zebrafish upon Aeromonas salmonicida and Staphylococcus aureus infection: striking similarities and obvious differences with mammals. Mol Immunol 44:295–301. doi:10.1016/j.molimm.2006.03.001
Luca F, Kashyap S, Southard C et al (2009) Adaptive variation regulates the expression of the human SGK1 gene in response to stress. PLoS Genet 5:e1000489. doi:10.1371/journal.pgen.1000489
Miller KM, Li S, Kaukinen KH et al (2011) Genomic signatures predict migration and spawning failure in wild Canadian salmon. Science 331:214–217. doi:10.1126/science.1196901
Miwata H, Yamada T, Okada M et al (1993) Serum amyloid A protein in acute viral infections. Arch Dis Child 68:210–214. doi:10.1136/adc.68.2.210
O’Reilly PT, Hamilton LC, McConnell SK, Wright JM (1996) Rapid analysis of genetic variation in Atlantic salmon (Salmo salar) by PCR multiplexing of dinucleotide and tetranucleotide microsatellites. Can J Fish Aquat Sci 53:2292–2298. doi:10.1139/f96-192
Oleksiak MF, Churchill GA, Crawford DL (2002) Variation in gene expression within and among natural populations. Nat Genet 32:261–266. doi:10.1038/ng983
Ouborg NJ, Pertoldi C, Loeschcke V et al (2010) Conservation genetics in transition to conservation genomics. Trends Genet 26:177–187. doi:10.1016/j.tig.2010.01.001
Paradis E (2010) Pegas: an R package for population genetics with an integrated-modular approach. Bioinformatics 26:419–420. doi:10.1093/bioinformatics/btp696
Parrish DL, Behnke RJ, Gephard SR et al (1998) Why aren’t there more Atlantic salmon (Salmo salar)? Can J Fish Aquat Sci 55:281–287. doi:10.1139/d98-012
Paterson S, Piertney SB, Knox D et al (2004) Characterization and PCR multiplexing of novel highly variable tetranucleotide Atlantic salmon (Salmo salar L.) microsatellites. Mol Ecol Notes 4:160–162. doi:10.1111/j.1471-8286.2004.00598.x
Pedersen KS, Kristensen TN, Loeschcke V (2005) Effects of inbreeding and rate of inbreeding in Drosophila melanogaster—Hsp70 expression and fitness. J Evol Biol 18:756–762. doi:10.1111/j.1420-9101.2005.00884.x
Reed DH, Frankham R (2001) How closely correlated are molecular and quantitative measures of genetic variation? a meta-analysis. Evolution 55:1095–1103. doi:10.1111/j.0014-3820.2001.tb00629.x
Roberge C, Einum S, Guderley H, Bernatchez L (2006) Rapid parallel evolutionary changes of gene transcription profiles in farmed Atlantic salmon. Mol Ecol 15:9–20. doi:10.1111/j.1365-294X.2005.02807.x
Roberge C, Guderley H, Bernatchez L (2007) Genomewide identification of genes under directional selection: gene transcription Q(ST) scan in diverging Atlantic salmon subpopulations. Genetics 177:1011–1022. doi:10.1534/genetics.107.073759
Roberge C, Normandeau E, Einum S et al (2008) Genetic consequences of interbreeding between farmed and wild Atlantic salmon: insights from the transcriptome. Mol Ecol 17:314–324. doi:10.1111/j.1365-294X.2007.03438.x
Rouillard J-M, Zuker M, Gulari E (2003) OligoArray 2.0: design of oligonucleotide probes for DNA microarrays using a thermodynamic approach. Nucleic Acids Res 31:3057–3062. doi:10.1093/nar/gkg426
Saeed AI, Sharov V, White J et al (2003) TM4: a free, open-source system for microarray data management and analysis. Biotechniques 34:374–378
Schlichting CD, Smith H (2002) Phenotypic plasticity: linking molecular mechanisms with evolutionary outcomes. Evol Ecol 16:189–211. doi:10.1023/A:1019624425971
Scott RJ, Poos MS, Noakes DLG, Beamish FWH (2005) Effects of exotic salmonids on juvenile Atlantic salmon behaviour. Ecol Freshw Fish 14:283–288. doi:10.1111/j.1600-0633.2005.00099.x
Smith EN, Kruglyak L (2008) Gene-environment interaction in yeast gene expression. PLoS Biol 6:e83. doi:10.1371/journal.pbio.0060083
Smyth GK (2005) limma: linear models for microarray data. In: Gentleman R, Carey VJ, Huber W et al (eds) Bioinforma Computational Biology Solution Using R Bioconductor. Springer, New York, pp 397–420
Spielman D, Brook BW, Briscoe DA, Frankham R (2004) Does inbreeding and loss of genetic diversity decrease disease resistance? Conserv Genet 5:439–448. doi:10.1023/B:COGE.0000041030.76598.cd
Storey JD, Madeoy J, Strout JL et al (2007) Gene-expression variation within and among human populations. Am J Hum Genet 80:502–509. doi:10.1086/512017
Townsend JP, Cavalieri D, Hartl DL (2003) Population genetic variation in genome-wide gene expression. Mol Biol Evol 20:955–963. doi:10.1093/molbev/msg106
Uno T, Ishizuka M, Itakura T (2012) Cytochrome P450 (CYP) in fish. Environ Toxicol Pharmacol 34:1–13. doi:10.1016/j.etap.2012.02.004
Van Zwol JA, Neff BD, Wilson CC (2012) The influence of non-native salmonids on circulating hormone concentrations in juvenile Atlantic salmon. Anim Behav 83:119–129. doi:10.1016/j.anbehav.2011.10.015
Vandersteen Tymchuk W, O’reilly P, Bittman J et al (2010) Conservation genomics of Atlantic salmon: variation in gene expression between and within regions of the Bay of Fundy. Mol Ecol 19:1842–1859. doi:10.1111/j.1365-294X.2010.04596.x
Wellband KW, Heath DD (2013) The relative contribution of drift and selection to transcriptional divergence among Babine Lake tributary populations of juvenile rainbow trout. J Evol Biol 26:2497–2508. doi:10.1111/jeb.12247
Whitehead A, Crawford DL (2006) Neutral and adaptive variation in gene expression. Proc Natl Acad Sci USA 103:5425–5430. doi:10.1073/pnas.0507648103
Yauk CL, Berndt ML, Williams A, Douglas GR (2004) Comprehensive comparison of six microarray technologies. Nucleic Acids Res 32:e124. doi:10.1093/nar/gnh123
Acknowledgments
This research was funded by Natural Sciences and Engineering Council of Canada and Ontario Ministry of Natural Resources. We thank W. Sloan, S. Ferguson, B. Lewis, A. Smith, C. Black, H. Dokter, J. Van Zwol, S. Garner, T. Hain, M.-C. Bellmare, M.-H. Greffard and H. Allegue for their assistance in semi-natural stream construction, fish feeding, sample collection and other support at the OMNR Codrington Research Facility. We thank R. Hepburn for microarray printing. We thank A. Kidd for microsatellite genotyping.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
He, X., Wilson, C.C., Wellband, K.W. et al. Transcriptional profiling of two Atlantic salmon strains: implications for reintroduction into Lake Ontario. Conserv Genet 16, 277–287 (2015). https://doi.org/10.1007/s10592-014-0657-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-014-0657-1