Abstract
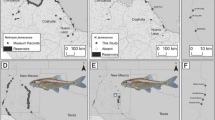
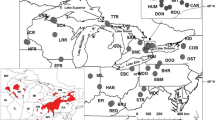
The Pugnose Shiner is a small minnow with a fragmented distribution across the Great Lakes and Upper Mississippi River in North America. The species is listed federally as endangered in Canada, and in the United States its status varies by state, from Special Concern to Endangered (as well as Extirpated). We conducted a thorough genetic assessment of the Pugnose Shiner using both microsatellite loci and mitochondrial DNA collected for samples across the species range. Our results indicate high levels of population differentiation suggesting restricted dispersal, in some cases at very small geographical scales. We also found strong evidence of small effective population sizes and one case of a genetic bottleneck. Although significant range-wide genetic variation was observed in both microsatellite loci and mitochondrial DNA, the species is best characterized as a single evolutionarily significant unit for conservation purposes.




Similar content being viewed by others
References
Abell R (2002) Conservation biology for the biodiversity crisis: a freshwater follow-up. Conserv Biol 16:1435–1437
Austin JD, Jelks HL, Tate B, Johnson AR, Jordan F (2011) Population genetic structure and conservation genetics of threatened Okaloosa darters (Etheostoma okaloosae). Conserv Genet 12:981–989
Bailey RM (1959) Distribution of the American cyprinid fish Notropis anogenus. Copeia 1959:119–123
Beaumont MA (1999) Detecting population expansion and decline using microsatellites. Genetics 153:2013–2029
Becker GC (1983) Fishes of Wisconsin. University of Wisconsin Press, Madison
Beneteau CL, Walter RP, Mandrak NE, Heath DD (2012) Range expansion by invasion: genetic characterization of invasion of the greenside darter (Etheostoma blennioides) at the northern edge of its distribution. Biol Invasions 14:191–201
Bernard AM, Ferguson MM, Noakes DLG, Morrison BJ, Wilson CC (2009) How different is different? Defining management and conservation units for a problematic exploited species. Can J Fish Aquat Sci 66:1617–1630
Bonin A, Bellemain E, Bronken Eidesen P, Pompanon C, Brochmann C, Taberlet P (2004) How to track and assess genotyping errors in population genetic studies. Mol Ecol 13:3261–3273
Broquet T, Angelone S, Jaquiery J, Joly P, Lena J-P, Lengagne T, Plenet S, Luquet E (2010) Genetic bottlenecks driven by population disconnection. Conserv Biol 24:1596–1605
Burridge CP, Gold JR (2003) Conservation genetic studies of the endangered Cape Fear shiner, Notropis mekistocholas (Teleostei: cyprinidae). Conserv Genet 4:219–225
Cavalli-Sforza LL, Edwards AWF (1967) Phylogenetic analysis: models and estimation procedures. Evolution 21:550–570
Chapuis M-P, Estoup A (2007) Microsatellite null alleles and estimation of population differentiation. Mol Biol Evol 24:621–631
Chikhi L, Sousa VC, Luisi P, Goossens B, Beaumont MA (2010) The confounding effects of population structure, genetic diversity and the sampling scheme on the detection and quantification of population size changes. Genetics 186:983–995
Cornuet J-M, Luikart G (1996) Description and power analysis of two tests for detecting recent population bottlenecks from allele frequency data. Genetics 144:2001–2014
COSEWIC (2013) COSEWIC assessment and update status report on the Pugnose Shiner Notropis anogenus in Canada. In: Committee on the Status of Endangered Wildlife in Canada.Ottawa
de Guia APO, Saitoh T (2007) The gap between the concept and definitions in the Evolutionarily Significant Unit: the need to integrate neutral genetic variation and adaptive variation. Ecol Res 22:604–612
DeHaan PW, Scheerer PD, Rhew R, Ardren WR (2012) Analyses of genetic variation in populations of Oregon Chub, a threatened floodplain minnow in a highly altered environment. Trans Am Fish Soc 141:533–549
DFO (2010) Recovery Potential Assessment of Pugnose Shiner (Notropis anogenus) in Canada. DFO Canadian Science Advisory Secretariat Science Advisory Report 2010/025
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Excoffier L, Lischer H (2010) Arlequin suite ver 35: a new series of programs to perform population genetics analyses under Linux and Windows. Mol Ecol Res 10:564–567
Faurby S, Pertoldi C (2012) The consequences of the unlikely but critical assumption of stepwise mutation in the population genetic software, MSVAR. Evol Ecol Res 14:859–879
Favé M-J, Turgeon J (2008) Patterns of genetic diversity in great lakes bloaters (Coregonus hoyi) with a view to future reintroduction in Lake Ontario. Conserv Genet 9:281–293
Fluker BL, Kuhajda BR, Lang NJ, Harris PM (2010) Low genetic diversity and small long-term population sizes in the spring endemic watercress darter, Etheostoma nuchale. Conserv Genet 11:2267–2279
Frankham R (2005) Genetics and Extinction. Biol Conserv 126:131–140
Frankham R, Ballou JD, Briscoe DA (2002) Introduction to Conservation Genetics. Cambridge University Press, Cambridge
Franklin I (1980) Evolutionary changes in small populations. In: Soule ME, Wilcox BA (eds) Conservation biology: an evolutionary-ecological perspective. Sinauer Associates, Sunderland, pp 135–140
Franklin IR, Frankham R (1998) How large must populations be to retain evolutionary potential? Anim Conserv 1:69–73
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Girod C, Vitalis R, Leblois R, Freville H (2011) Inferring population decline and expansion from microsatellite data: a simulation-based evaluation of the Msvar method. Genetics 188:165–179
Gold JR, Saillant E, Burridge CP, Blanchard A, Patton JC (2004) Population structure and effective size in critically endangered Cape Fear shiners Notropis mekistocholas. Southeast Nat 3:89–102
Goudet J (1995) FSTAT (Version 12): a computer program to calculate F-statistics. J Hered 86:485–486
Green DM (2005) Designatable units for status assessment of endangered species. Conserv Biol 19:1813–1820
Holm E, Mandrak NE (2002) COSEWIC status report on the pugnose shiner Notropis anogenus in Canada: In Committee on the status of endangered wildlife in Canada, pp 1-15
Hubisz MJ, Falush D, Stephens M, Pritchard JK (2009) Inferring weak population structure with the assistance of sample group information. Mol Ecol Res 9:1322–1332
Ingvarsson PK (2001) Restoration of genetic variation lost- the genetic rescue hypothesis. Trends Ecol Evol 16:62–63
IUCN (2013) IUCN Red List of Threatened Species. Version 2013.1. http://www.iucnredlist.org. Accessed 21 Oct 2013
Jelks HL, Walsh SJ, Burkhead NM, Contreras-Balderas S, Diaz-Pardo E, Hendrickson DA, Lyons J, Mandrak N, McCormick F, Platania SP, Porter BA, Renaud CB, Schmitter-Soto JJ, Taylor EB, Warren MLJ (2008) Conservation status of imperiled North American freshwater and diadromous fishes. Fisheries 33:372–407
Langella O (1999) Populations: a population genetic software, CNRS UPR9034. http://bioinformatics.org/~tryphon/populations/. Accessed 17 May 2012
McCraney WT, Goldsmith G, Jacobs DK, Kinziger AP (2010) Rampant drift in artificially fragmented populations of the endangered tidewater goby (Eucyclogobius newberryi). Mol Ecol 19:3315–3327
Miller MP (1997) Tools for population genetic analyses (TFPGA) v 13 A windows program for the analysis of allozyme and molecular population genetic data. http://www.marksgeneticsoftware.net/. Accessed 12 May 2012
Moritz C (1994) Defining ‘ Evolutionarily Significant Units’ for conservation. TREE 9:373–375
NatureServe (2013) NatureServe Explorer: an online encyclopedia of life [web application]. Version 7.1. NatureServe, Arlington, Virginia. http://www.natureserve.org/explorer. Accessed 21 Oct 2013
Page LM, Burr BM (2011) A field guide to freshwater fishes of North America north of Mexico. The Peterson Field Guide Series, 2nd ed edn. Houghton Mifflin Co, Boston
Peery MZ, Kirby R, Reid BN, Stoelting R, Douct-Beer E, Robinson S, Vasquez-Carrillo C, Pauli JN, Palsboll PJ (2012) Reliability of genetic bottleneck tests for detecting recent population declines. Mol Ecol 21:3403–3418
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Rambaut A, Drummond AJ (2009) Tracer v1.5. http://tree.bio.ed.ac.uk/software/tracer/. Accessed 17 May 2012
Raymond M, Rousset F (1995) GENEPOP (version 12): population genetics software for exact tests and ecumenicism. J Hered 86:248–249
Renshaw M, Carson E, Hanna A, Rexroad CEI, Krabbenhoft T, Gold J (2009) Microsatellite markers for species of the genus Dionda (Cyprinidae) from the American southwest. Conserv Genet 10:1569–1575
Robinson JD, Simmons JW, Williams AS, Moyer GR (2013) Population structure and genetic diversity in the endangered Bluemask Darter (Etheostoma akatulo). Conserv Genet 14:79–92
Rosenberg NA (2004) DISTRUCT: a program for the graphical display of population structure. Mol Ecol Notes 4:137–138
Rosenberg NA, Pritchard JK, Weber JL, Cann HM, Kidd KK, Zhivotovsky LA, Feldman MW (2002) Genetic structure of human populations. Science 298:2381–2385
Schonhuth S, Doadrio I (2003) Phylogenetic relationships of Mexican minnows of the genus Notropis (Actinopterygii, Cyprinidae). Biol J Linn Soc 80:323–337
Sepulveda-Villet OJ, Stepien CA (2011) Fine-scale population genetic structure of the yellow perch Perca flavescens in Lake Erie. Can J Fish Aquat Sci 68:1435–1453
Sousa VC, Penha F, Pala I, Chikhi L, Coelho MM (2010) Conservation genetics of a critically endangered Iberian minnow: evidence of population decline and extirpations. Anim Conserv 13:162–171
Spielman D, Brook BW, Frankham R (2004) Most species are not driven to extinction before genetic factors impact them. Proc Nat Acad Sci 101:15261–15264
Stepien CA, Murphy DJ, Strange RM (2007) Broad.to fine-scale population genetic patterning in the smallmouth bass Micropterus dolomieu across the Laurentian Great Lakes and beyond: an interplay of behaviour and geography. Mol Ecol 16:1605–1624
Stepien CA, Murphy DJ, Lohner RN, Sepulveda-Villet OJ, Haponski AE (2009) Signatures of vicariance, postglacial dispersal and spawning philopatry: population genetics of the walleye Sander vitreus. Mol Ecol 18:3411–3428
Sterling KA, Reed DH, Noonan BP, Warren ML Jr (2012) Genetic effects of habitat fragmentation and population isolation on Etheostoma raneyi (Percidae). Conserv Genet 13:859–872
Storz JF, Beamont MA (2002) Testing for genetic evidence of population expansion and contraction: an empirical analysis of microsatellite DNA variation using a hierarchical Bayesian model. Evolution 56:154–166
Takezaki N, Nei M (1996) Genetic distances and reconstruction of phylogenetic trees from microsatellite DNA. Genetics 144:389–399
Underhill JC (1957) The distribution of Minnesota minnows and darters in relation to Pleistocene glaciation. Minn Mus Nat Hist Occ Pap 7:1–45
Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P (2004) MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Mol Ecol Notes 4:535–538
Vila C, Sundqvist A-K, Flagstad O, Seddon J, Bjornerfeldt S, Kojola I, Casulli A, Sand H, Wabakken P, Ellegren H (2003) Rescue of a severely bottlenecked wolf (Canis lupus) population by a single immigrant. Proc Royal Soc London B 270:91–97
Waples RS (1991) Pacific Salmon, Oncorhynchus spp., and the definition of “Species” under the Endangered Species Act. Mar Fish Rev 53:11–22
Waples RS (1998) Evolutionarily significant units, distinct population segments, and the Endangered Species Act: reply to Pennock and Dimmick. Conserv Bio 12:718–721
Waples RS, Do C (2008) LDNE: a program for estimating effective population size from data on linkage disequilibrium. Mol Ecol Res 8:753–756
Waples RS, Do C (2010) Linkage disequilibrium estimates of contemporary Ne using highly variable genetic markers: a largely untapped resource for applied conservation and evolution. Evol App 3:244–262
Waples RS, Gaggiotti O (2006) What is a population? An empirical evaluation of some genetic methods for identifying the number of gene pools and their degree of connectivity. Mol Ecol 15:1419–1439
Whitlock MC, McCauley DE (1999) Indirect measures of gene flow and migration: F ST ≠ 1/(4Nm + 1). Heredity 82:117–125
Willi Y, Van Buskirk J, Hoffman AA (2006) Limits to the adaptive potential of small populations. Annu Rev Ecol Sys 37:433–458
Willi Y, Griffin P, Van Buskirk J (2013) Drift load in populations of small size and low density. Hered 110:296–302
Acknowledgments
Funding for this study was provided by the Species at Risk program of Fisheries and Oceans Canada (DFO), World Wildlife Fund (WWF-Canada), Ontario Ministry of Natural Resources (OMNR), and an NSERC Discovery Grant to NRL. M.R. McCusker was supported by the NSERC Visiting Fellowship program. We are grateful for sampling assistance from DFO field crews, Doug Carlson, Ross Abbett, Kristen Brochu, Devin Bloom, Konrad Schmidt, Kimberly Strand, John Lyons, Michael Hawkins, and Edward Thelen. We would also like to thank all anonymous reviewers for their constructive comments.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
McCusker, M.R., Mandrak, N.E., Egeh, B. et al. Population structure and conservation genetic assessment of the endangered Pugnose Shiner, Notropis anogenus . Conserv Genet 15, 343–353 (2014). https://doi.org/10.1007/s10592-013-0542-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-013-0542-3