Abstract
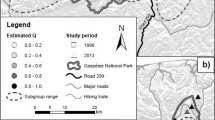
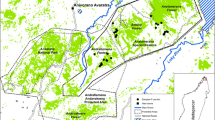
Endangered species worldwide exist in remnant populations, often within fragmented landscapes. Although assessment of genetic diversity in fragmented habitats is very important for conservation purposes, it is usually impossible to evaluate the amount of diversity that has actually been lost. Here, we compared population structure and levels of genetic diversity within populations of spotted suslik Spermophilus suslicus, inhabiting two different parts of the species range characterized by different levels of habitat connectivity. We used microsatellites to analyze 10 critically endangered populations located at the western part of the range, where suslik habitat have been severely devastated due to agriculture industrialization. Their genetic composition was compared with four populations from the eastern part of the range where the species still occupies habitat with reasonable levels of connectivity. In the western region, we detected extreme population structure (F ST = 0.20) and levels of genetic diversity (Allelic richness ranged from 1.45 to 3.07) characteristic for highly endangered populations. Alternatively, in the eastern region we found significantly higher allelic richness (from 5.09 to 5.81) and insignificant population structure (F ST = 0.03). As we identified a strong correlation between genetic and geographic distance and a lack of private alleles in the western region, we conclude that extreme population structure and lower genetic diversity is due to recent habitat loss. Results from this study provide guidelines for conservation and management of this highly endangered species.


Similar content being viewed by others
References
Amos W, Balmford A (2001) When does conservation genetics matter? Heredity 87:257–265
Barnett R, Yamaguchi N, Barnes I, Cooper A (2006) Lost populations and preserving genetic diversity in the lion Panthera leo: implications for its ex situ conservation. Conserv Genet 7:507–514
Berthier K, Charbonnel M, Galan M, Chaval Y, Cosson J-F (2006) Migration and recovery of the genetic diversity during the increasing density phase in cyclic vole populations. Mol Ecol 15:2665–2676
Chapuisat M, Goudet J, Keller L (1997) Microsatellites reveal high population viscosity and limited dispersal in ant Formica paralugubris. Evolution 51:475–482
Ciofi C, Bruford MW (1999) Genetic structure and gene flow among Komodo dragon populations inferred by microsatellite loci analysis. Mol Ecol 8:17–30
Di Renzo A, Peterson AC, Garza JC, Valdes AM, Slatkin M, Freimer NB (1994) Mutational processes of simple-sequence repeat loci in human populations. Proc Natl Acad Sci USA 91:3166–3170
Excoffier L, Smouse P, Quattro J (1992) Analysis of molecular variance inferred from genetic distances among DNA haplotypes: application to human mitochondrial DNA restriction data. Genetics 131:479–491
Frankel OH, Soule ME (1981) Conservation and evolution. Cambridge University Press, New York, New York
Frankham R, Ballou JD, Briscoe DA (2002) Conservation genetics. Cambridge University Press, Cambridge
Garner A, Rachlow JL, Waits LP (2005) Genetic diversity and population divergence in fragmented habitats: conservation of Idaho ground squirrels. Conserv Genet 6:759–774
Garza JC, Williamson EG (2001) Detection of reduction in population size using data from microsatellite loci. Mol Ecol 10:305–318
Głowaciński Z, Męczyński S (2001) Suseł perełkowany Spermophilus suslicus. In: Głowaciński (ed) Polish red data book of animals (Vertebrates). Państwowe wydawnictwa leśne i rolnicze. Warszawa, Poland
Gondek A, Verduijn M, Wolff K (2006) Polymorphic microsatellite markers for endangered spotted suslik, Spermophilus suslicus. Mol Ecol Notes 6:359–361
Gondek A (2004) Sytuacja susła perełkowanego w Polsce—zagrożenia i program ochrony. Biuletyn Monitoringu Przyrody 1/2004:21–27
Goudet J (2002) FSTAT (v. 1.2): a computer program to calculate F-statistics. J Heredity 86:485–486
Gyllenberg M, Hanski I (1992) Single-species metapopulation dynamics: a structured model. Theor Pop Biol 42:35–61
Hanslik S, Kruckenhauser L (2000) Microsatellite loci for two European sciurid species (Marmota marmota, Spermophilus citellus). Mol Ecol 9:2163–2165
Hazlitt SL, Goldizen AW, Eldridge MDB (2006) Significant patterns of population genetic structure and limited gene flow in a threatened macropodid marsupial despite continuous habitat in southeast Queensland, Australia. Conserv Genet 7:675–689
Hirota T, Hirohata T, Hiroshi M, Toshiyuki S, Yoshiaki O (2004) Population structure of the large Japanese field mouse, Apodemus speciosus (Rodentia: Muridae), in suburban landscape, based on mitochondrial D-loop sequences. Mol Ecol 13:3275–3282
Honnay LO Adriaens D, Coart E, Jacquemyn H, Roldan-Ruiz I (2007) Genetic diversity within and between remnant populations of the endangered calcareous grassland plant Globularia bisnagarica. Conserv Genet 8:293–303
Kalinowski ST (2005) HP-RARE 1.0: a computer program for performing rarefaction on measures of allelic richness. Mol Ecol Notes 5:187–189
Lobkov VA (1999) Spotted suslik of the north-western region by the Black Sea: biology and regulation of it’s population. AstroPrint, Odessa, Ukraine
Lowe AJ, Jourde B, Breyne P, Colpaert N, Navarro C, Wilson J, Cavers S (2003) Fine-scale genetic structure and gene flow within Costa Rican populations of mahogany (Swietenia macrophylla). Heredity 90:268–275
MacAvoy ES, McGibbon LM, Sainsbury JP, Lawrence H, Wilson CA, Daugherty CH, Chambers GK (2007) Genetic variation in island populations of tuatara (Sphenodon spp) inferred from microsatellite markers. Conserv Genet 8:305–318
Mantel N (1967) The detection of disease clustering and a generalized regression approach. Cancer Res 27:209–220
Maruyama T, Fuerst PA (1985) Population bottlenecks and non-equilibrium models in population genetics. II. Number of alleles in a small population that was formed by a recent bottleneck. Genetics 111:675–689
May B, Gavin TA, Sherman PW, Korves TM (1997) Characterization of microsatellite loci in the northern Idaho ground squirrel Spermophilus brunneus brunneus. Mol Ecol 6:399–400
Męczyński S (1991) Występowanie susła perełkowanego Spermophilus suslicus w Polsce i koncepcje jego ochrony. Ochrona przyrody 48:207–237
Męczyński S, Grądziel T, Styka R, Duda P, Próchnicki K (2001) Inwentaryzacja stanowisk susła perełkowanego na Zamojszczyźnie w roku 2001. Spotted suslik monitoring evaluation- a raport for Lublin Voivodeship Nature Conservator, Lublin, Poland
Nei M, Maruyama T, Chakraborty R (1975) The bottleneck effect and genetic variability in populations. Evolution 29:1–10
Nei M 1978: Estimation of average heterozygosity and genetic distance from a number of individuals. Genetics 89:538–590
Pamilo P (1985) Effect of inbreeding on genetic relatedness. Hereditas 103:195–200
Piry S, Luikart G, Cornuet J-M (1999) BOTTLENECK: a computer program for detecting recent reductions in the effective population size using allele frequency data. J Heredity 95:536–539
Pritchard JK, Stevens M, Donnely P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Queller DC, Goodnight KF (1989) Estimating relatedness using genetic markers. Evolution 42:258–275
Ramirez O, Altet L, Ensen C, Vila C, Sanchez A, Ruiz A (2006) Genetic assessment of the Iberian wolf Canis lupus signatus captive breeding program. Conserv Genet 7:861–878
Rosenbaum PA, Robertson JM, Zamudio KR (2007) Unexpectedly low genetic divergences among populations of the threatened bog turtle (Glyptemys muhlenbergii). Conserv Genet 8:331–342
Rousset F, (1997) Genetic differentiation and estimation of gene flow from F-statistics under isolation by distance. Genetics 145:1219–1228
Schneider S, Roessli D, Excoffier L (2000): ARLEQUIN ver. 2.000: a software for population genetics data analysis. Genetics and Biometry Laboratory, University of Geneva, Switzerland
Schwartz MK, Mills LS, Ortega Y, Ruggiero LF, Allendorf FW (2003) Landscape location affects genetic variation of Canada lynx (Lynx canadensis). Mol Ecol 12:1807–1816
Slatkin M (1985) Rare alleles as indication of gene flow. Evolution 39:53–65
Slatkin M (1995) A measure of population subdivision based on microsatellite allele frequencies. Genetics 139:457–462
Surdacki S (1963) Zmiany w rozmieszczeniu i liczebności Citellus suslicus na Lubelszczyźnie w okresie 1954–1961. Acta Theriol 7:79–89
Titov SV (2003) Juvenile dispersal in the colonies of Spermophilus major and S. suslicus ground squirrels. Russ J Ecol 34:255–260
Vucetich JA, Waite TA (2003) Spatial patterns of demography and genetic processes across the species range: null hypotheses for landscape conservation genetics. Conserv Genet 4:639–645
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population structure. Evolution 38:1358–1370
Acknowledgements
This research was funded by the Polish State Committee for Scientific Research (grant no.: 3P04G00924) and the Polish Ministry of Science (grant no.: N30309031/2938).
We want to acknowledge Dr Vlodimir Lobkov from the Odessa National University, Ukraine for providing samples from the eastern region and to all the people who helped us to collect samples from the western region. We are also grateful to the Association of Zamość Natural Landscape Parks and the Lublin Voivodeship Nature Conservator for their support in our research.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Biedrzycka, A., Konopiński, M.K. Genetic variability and the effect of habitat fragmentation in spotted suslik Spermophilus suslicus populations from two different regions. Conserv Genet 9, 1211–1221 (2008). https://doi.org/10.1007/s10592-007-9442-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10592-007-9442-8