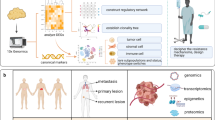
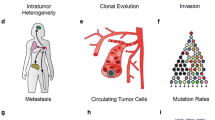
Abstract
Single-cell sequencing technologies have undergone rapid development and adoption by the scientific community in the past 5 years, fueling discoveries about the etiology, pathogenesis, and treatment responsiveness of individual tumor cells within cancer ecosystems. Most of the advancements in our understanding of cancer with these new technologies have focused on basic tumor biology. However, the knowledge produced by these and other studies are beginning to provide biomarkers and drug targets for clinically-relevant subpopulations within a tumor, creating opportunities for the development of biologically-informed, clone-specific combination treatment strategies. Here we provide an overview of the development of the field of single-cell cancer sequencing and provide a roadmap for shepherding these technologies from research tools to diagnostic instruments that provide high-resolution, treatment-directing details of tumors to clinical oncologists.


Similar content being viewed by others
Abbreviations
- TCGA:
-
The cancer genome atlas
- ICGC:
-
International cancer genome consortium
- TARGET:
-
Therapeutically applicable research to generate effective treatments
- PCR:
-
Polymerase chain reaction
- scRNA-seq:
-
Single cell RNA sequencing
- MDA:
-
Multiple displacement amplification
- ATAC-Seq:
-
Assay for transposase-accessible chromatin with high-throughput sequencing
References
Lander ES et al (2001) Initial sequencing and analysis of the human genome. Nature 409(6822):860–921
Tucker T, Marra M, Friedman JM (2009) Massively parallel sequencing: the next big thing in genetic medicine. Am J Hum Genet 85(2):142–154
Consortium IT (2020) Pan-cancer analysis of whole genomes. Nature 578(7793):82–93
Ma X et al (2018) Pan-cancer genome and transcriptome analyses of 1,699 paediatric leukaemias and solid tumours. Nature 555(7696):371–376
Presley CJ et al (2018) Association of broad-based genomic sequencing with survival among patients with advanced non-small cell lung cancer in the community oncology setting. JAMA 320(5):469–477
Rosenthal R et al (2017) Deciphering genetic intratumor heterogeneity and its impact on cancer evolution. Annu Rev Cancer Biol 1:223–240
Dagogo-Jack I, Shaw AT (2018) Tumour heterogeneity and resistance to cancer therapies. Nat Rev Clin Oncol 15(2):81–94
Lee D, Park Y, Kim S (2021) Towards multi-omics characterization of tumor heterogeneity: a comprehensive review of statistical and machine learning approaches. Brief Bioinform 22(3):bbaa188
Zhang Q et al (2019) Integrated multiomic analysis reveals comprehensive tumour heterogeneity and novel immunophenotypic classification in hepatocellular carcinomas. Gut. https://doi.org/10.1136/gutjnl-2019-318912
Liu J, Dang H, Wang XW (2018) The significance of intertumor and intratumor heterogeneity in liver cancer. Exp Mol Med 50(1):e416–e416
Stuart T, Satija R (2019) Integrative single-cell analysis. Nat Rev Genet 20(5):257–272
Linnarsson S, Teichmann SA (2016) Single-cell genomics: coming of age. Springer, Berlin
Gawad C, Koh W, Quake SR (2016) Single-cell genome sequencing: current state of the science. Nat Rev Genet 17(3):175–188
Luquette LJ et al (2019) Identification of somatic mutations in single cell DNA-seq using a spatial model of allelic imbalance. Nat Commun 10(1):1–14
Sokolenko AP, Imyanitov EN (2018) Molecular diagnostics in clinical oncology. Front Mol Biosci 5:76
Tang X et al (2019) The single-cell sequencing: new developments and medical applications. Cell Biosci 9(1):1–9
Klein AM et al (2015) Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161(5):1187–1201
Macosko EZ et al (2015) Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161(5):1202–1214
Picelli S et al (2014) Tn5 transposase and tagmentation procedures for massively scaled sequencing projects. Genome Res 24(12):2033–2040
Pellegrino M et al (2018) High-throughput single-cell DNA sequencing of acute myeloid leukemia tumors with droplet microfluidics. Genome Res 28(9):1345–1352
Huang L et al (2015) Single-cell whole-genome amplification and sequencing: methodology and applications. Annu Rev Genomics Hum Genet 16:79–102
Dean FB et al (2002) Comprehensive human genome amplification using multiple displacement amplification. Proc Natl Acad Sci USA 99(8):5261–5266
Long N et al (2020) Recent advances and application in whole-genome multiple displacement amplification. Quant Biol 8:1–16
Gonzalez-Pena V et al (2021) Accurate genomic variant detection in single cells with primary template-directed amplification. Proc Natl Acad Sci USA. https://doi.org/10.1073/pnas.2024176118
Allis CD, Jenuwein T (2016) The molecular hallmarks of epigenetic control. Nat Rev Genet 17(8):487–500
Virani S et al (2012) Cancer epigenetics: a brief review. ILAR J 53(3–4):359–369
Buenrostro JD et al (2015) ATAC-seq: a method for assaying chromatin accessibility genome‐wide. Curr Protoc Mol Biol 109(1):21. 1-21.29. 9.
Li Y, Tollefsbol TO (2011) DNA methylation detection: bisulfite genomic sequencing analysis. In: Epigenetics protocols. Springer, Berlin, pp 11–21
Kashima Y et al (2020) Single-cell sequencing techniques from individual to multiomics analyses. Exp Mol Med 52(9):1419–1427
Robinson WH (2015) Sequencing the functional antibody repertoire—diagnostic and therapeutic discovery. Nat Rev Rheumatol 11(3):171–182
Van der Auwera GA, O’Connor BD (2020) Genomics in the cloud: using Docker, GATK, and WDL in Terra. O’Reilly Media, Sebastopol
Hwang B, Lee JH, Bang D (2018) Single-cell RNA sequencing technologies and bioinformatics pipelines. Exp Mol Med 50(8):96
Laird PW (2010) Principles and challenges of genome-wide DNA methylation analysis. Nat Rev Genet 11(3):191–203
Guo H et al (2013) Single-cell methylome landscapes of mouse embryonic stem cells and early embryos analyzed using reduced representation bisulfite sequencing. Genome Res 23(12):2126–2135
Melchor L et al (2014) Single-cell genetic analysis reveals the composition of initiating clones and phylogenetic patterns of branching and parallel evolution in myeloma. Leukemia 28(8):1705–1715
Zugazagoitia J et al (2016) Current challenges in cancer treatment. Clin Ther 38(7):1551–1566
Waldman AD, Fritz JM, Lenardo MJ (2020) A guide to cancer immunotherapy: from T cell basic science to clinical practice. Nat Rev Immunol 20(11):651–668
Casasent AK et al (2018) Multiclonal invasion in breast tumors identified by topographic single cell sequencing. Cell 172(1–2):205-217e12
Waylen LN et al (2020) From whole-mount to single-cell spatial assessment of gene expression in 3D. Commun Biol 3(1):1–11
Gawad C, Koh W, Quake SR (2014) Dissecting the clonal origins of childhood acute lymphoblastic leukemia by single-cell genomics. Proc Natl Acad Sci USA 111(50):17947–52
Lawson DA et al (2018) Tumour heterogeneity and metastasis at single-cell resolution. Nat Cell Biol 20(12):1349–1360
Metzker ML (2010) Sequencing technologies—the next generation. Nat Rev Genet 11(1):31–46
Funding
C.G.’s work on single-cell cancer sequencing is supported by the Chan Zuckerberg Biohub Investigator Program, a Burroughs Wellcome Fund Career Award for Medical Scientists, and an NIH Director’s New Innovator Award (1DP2CA239145).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Xia, Y., Gawad, C. Bringing precision oncology to cellular resolution with single-cell genomics. Clin Exp Metastasis 39, 79–83 (2022). https://doi.org/10.1007/s10585-021-10129-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10585-021-10129-4