Abstract
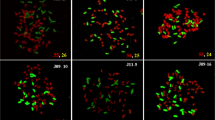
Modern sugarcane cultivars are derived from the hybridization of Saccharum officinarum (2n = 80) and S. spontaneum (2n = 40–128), leading to a variety of complex genomes with highly polyploid and varied chromosome structures. These complex genomes have hindered deciphering the genome structure and marker-assisted selection in sugarcane breeding. Ten cultivars were analyzed by fluorescence in situ hybridization adopting chromosome painting and S. spontaneum–specific probes. The results showed six types of chromosomes in the studied cultivars, including S. spontaneum or S. officinarum chromosomes, interspecific recombinations from homoeologous or nonhomoeologous chromosomes, and translocations of S. spontaneum or S. officinarum chromosomes. The results showed unexpectedly high proportions of interspecific recombinations in these cultivars (11.9–40.9%), which renew our knowledge that less than 13% of chromosomes result from interspecific exchanges. Also, the results showed a high frequency of translocations (an average of 2.15 translocations per chromosome) between S. officinarum chromosomes. The diverse types of chromosomes in cultivars imply that hybrid gametes of S. spontaneum and S. officinarum may form unusual chromosome pairs, including homoeologous or nonhomoeologous chromosomes either between or within S. spontaneum and S. officinarum. Moreover, we consistently observed 11 or 12 copies for the four studied chromosomes, i.e., chromosomes 1, 2, 7, and 8, suggesting steady transmission during the breeding program. By comparison, we found a relatively fewer copies of S. spontaneum chromosome 1 than those of S. spontaneum chromosomes 2, 7, and 8. These results provide deep insights into the structure of cultivars and may facilitate chromosome-assisted selection in sugarcane breeding.




Similar content being viewed by others
Availability of data and material
Not applicable.
Change history
07 April 2022
A Correction to this paper has been published: https://doi.org/10.1007/s10577-022-09690-9
Abbreviations
- DAPI:
-
4ʹ,6ʹ-Diamidino-phenylindole
- FISH:
-
Fluorescence in situ hybridization
- Mb:
-
Megabase
References
Albert PS, Zhang T, Semrau K, Rouillard JM, Kao YH, Wang CR, Danilova TV, Jiang J, Birchler JA (2019) Whole-chromosome paints in maize reveal rearrangements, nuclear domains, and chromosomal relationships. Proc Natl Acad Sci U S A 116:1679–1685
Bi Y, Zhao Q, Yan W, Li M, Liu Y, Cheng C, Zhang L, Yu X, Li J, Qian C, Wu Y, Chen J, Lou Q (2020) Flexible chromosome painting based on multiplex PCR of oligonucleotides and its application for comparative chromosome analyses in Cucumis. Plant J 102:178–186
Bremer G (1961) Problems in breeding and cytology of sugar cane. Euphytica 10:59–78
Chen Y, Wang Y, Wang K, Zhu X, Guo W, Zhang T, Zhou B (2014) Construction of a complete set of alien chromosome addition lines from Gossypium australe in Gossypium hirsutum: morphological, cytological, and genotypic characterization. Theor Appl Genet 127:1105–1121
Cuadrado A, Acevedo R, Díaz M, de la Espina S, Jouve N, de la Torre C (2004) Genome remodelling in three modern S. officinarum × S. spontaneum sugarcane cultivars. J Exp Bot 55:847–854
D’Hont A, Grivet L, Feldmann P, Glaszmann JC, Rao S, Berding N (1996) Characterisation of the double genome structure of modern sugarcane cultivars (Saccharum spp.) by molecular cytogenetics. Mol Gen Genet MGG 250:405–413
D’Hont A, Ison D, Alix K, Roux C, Glaszmann JC (1998) Determination of basic chromosome numbers in the genus Saccharum by physical mapping of ribosomal RNA genes. Genome 41:221–225
de Setta N, Monteiro-Vitorello CB, Metcalfe CJ, Cruz GMQ, Del Bem LE, Vicentini R, Nogueira FTS, Campos RA, Nunes SL, Turrini PCG, Vieira AP, Ochoa Cruz EA, Corrêa TCS, Hotta CT, de Mello VA, Vautrin S, da Trindade AS, de Mendonça VM, Lembke CG, Sato PM, de Andrade RF, Nishiyama MY, Cardoso-Silva CB, Scortecci KC, Garcia AAF, Carneiro MS, Kim C, Paterson AH, Bergès H, D’Hont A, de Souza AP, Souza GM, Vincentz M, Kitajima JP, Van Sluys M-A (2014) Building the sugarcane genome for biotechnology and identifying evolutionary trends. BMC Genomics 15:540
Garsmeur O, Droc G, Antonise R, Grimwood J, Potier B, Aitken K, Jenkins J, Martin G, Charron C, Hervouet C, Costet L, Yahiaoui N, Healey A, Sims D, Cherukuri Y, Sreedasyam A, Kilian A, Chan A, Van Sluys MA, Swaminathan K, Town C, Berges H, Simmons B, Glaszmann JC, van der Vossen E, Henry R, Schmutz J, D’Hont A (2018) A mosaic monoploid reference sequence for the highly complex genome of sugarcane. Nat Commun 9:2638
Heinz D (1987) Sugarcane improvement through breeding. Elsevier, Amsterdam
Hu ZH (2018) Analysis on the production situation and development trend of sugarcane production in China. Sugarcane Canesugar 47:58–66
Huang Y, Chen H, Han J, Zhang Y, Ma S, Yu G, Wang Z, Wang K (2020) Species-specific abundant retrotransposons elucidate the genomic composition of modern sugarcane cultivars. Chromosoma 129:45–55
Huang Y, Ding W, Zhang M, Han J, Jing Y, Yao W, Hasterok R, Wang Z, Wang K (2021) The formation and evolution of centromeric satellite repeats in Saccharum species. Plant J 106:616–629
Irvine JE (1999) Saccharum species as horticultural classes. Theor Appl Genet 98:186–194
Meng Z, Zhang Z, Yan T, Lin Q, Wang Y, Huang W, Huang Y, Li Z, Yu Q, Wang J, Wang K (2018) Comprehensively characterizing the cytological features of Saccharum spontaneum by the development of a complete set of chromosome-specific oligo probes. Front Plant Sci 9:1624
Meng Z, Han J, Lin Y, Zhao Y, Lin Q, Ma X, Wang J, Zhang M, Zhang L, Yang Q, Wang K (2020) Characterization of a Saccharum spontaneum with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theor Appl Genet 133:187–199
Meng Z, Wang Q, Khurshid H, Raza G, Han J, Wang B, Wang K (2021) Chromosome painting provides insights into the genome structure and evolution of sugarcane. Front Plant Sci 12:731664
Nagaki K, Tsujimoto H, Sasakuma T (1998) A novel repetitive sequence of sugar cane, SCEN family, locating on centromeric regions. Chromosome Res 6:295–302
Panje RR, Babu CN (1960) Studies in Saccharum spontaneum distribution and geographical association of chromosome numbers. Cytologia 25:152–172
Piperidis N, D’Hont A (2020) Sugarcane genome architecture decrypted with chromosome specific oligo probes. Plant J
Piperidis G, Piperidis N, D’Hont A (2010) Molecular cytogenetic investigation of chromosome composition and transmission in sugarcane. Mol Genet Genomics 284:65–73
Price S (1963) Cytogenetics of modern sugar canes. Econ Bot 17:97–106
Rines HW, Phillips RL, Kynast RG, Okagaki RJ, Galatowitsch MW, Huettl PA, Stec AO, Jacobs MS, Suresh J, Porter HL, Walch MD, Cabral CB (2009) Addition of individual chromosomes of maize inbreds B73 and Mo17 to oat cultivars Starter and Sun II: maize chromosome retention, transmission, and plant phenotype. Theor Appl Genet 119:1255–1264
Roach BT (1969) Cytological studies in Saccharum chromosome transmission in inter-specific and inter-generic crosses. Proc Int Soc Sugar Cane Technol 13:901–920
Sreenivasan TV (1975) Cytogenetical studies in Saccharum spontaneum. Proc Indian Acad Sci 81:131–144
Sreenivasan TV, Jagathesan D (1975) Meiotic abnormalities in Saccharum spontaneum. Euphytica 24:543–549
Vieira MLC, Almeida CB, Oliveira CA, Tacuatia LO, Munhoz CF, Cauz-Santos LA, Pinto LR, Monteiro-Vitorello CB, Xavier MA, Forni-Martins ER (2018) Revisiting meiosis in sugarcane: chromosomal irregularities and the prevalence of bivalent configurations. Front Genet 9:213
Zhang W, Zuo S, Li Z, Meng Z, Han J, Song J, Pan YB, Wang K (2017) Isolation and characterization of centromeric repetitive DNA sequences in Saccharum spontaneum. Sci Rep 7:41659
Zhang J, Zhang X, Tang H, Zhang Q, Hua X, Ma X, Zhu F, Jones T, Zhu X, Bowers J, Wai CM, Zheng C, Shi Y, Chen S, Xu X, Yue J, Nelson DR, Huang L, Li Z, Xu H, Zhou D, Wang Y, Hu W, Lin J, Deng Y, Pandey N, Mancini M, Zerpa D, Nguyen JK, Wang L, Yu L, Xin Y, Ge L, Arro J, Han JO, Chakrabarty S, Pushko M, Zhang W, Ma Y, Ma P, Lv M, Chen F, Zheng G, Xu J, Yang Z, Deng F, Chen X, Liao Z, Zhang X, Lin Z, Lin H, Yan H, Kuang Z, Zhong W, Liang P, Wang G, Yuan Y, Shi J, Hou J, Lin J, Jin J, Cao P, Shen Q, Jiang Q, Zhou P, Ma Y, Zhang X, Xu R, Liu J, Zhou Y, Jia H, Ma Q, Qi R, Zhang Z, Fang J, Fang H, Song J, Wang M, Dong G, Wang G, Chen Z, Ma T, Liu H, Dhungana SR, Huss SE, Yang X, Sharma A, Trujillo JH, Martinez MC, Hudson M, Riascos JJ, Schuler M, Chen LQ, Braun DM, Li L, Yu Q, Wang J, Wang K, Schatz MC, Heckerman D, Van Sluys MA, Souza GM, Moore PH, Sankoff D, VanBuren R, Paterson AH, Nagai C, Ming R (2018) Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet 50:1565–1573
Acknowledgements
We would like to thank the National Field Genebank of Sugarcane Germplasm of China and National Infrastructure for Crop Germplasm Resources--Sugarcane platform of China for supplying us the S. spontaneum plants. We thank Zhuang Meng for the design of oligos and technical assistance in cytogenetic assay.
Funding
This work was supported by the National Natural Science Foundation of China (31771862), the Startup Foundation from Nantong University (03083074), Science and Technology Innovation Fund of Fujian Agricultural and Forestry University (CXZX2020001A), the Research Program of Guangxi Key Laboratory for Sugarcane Biology (GXKLSCB-20190203), and State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources (SKLCUSA-b201808).
Author information
Authors and Affiliations
Contributions
KW and BW designed the research and drafted the manuscript. JL, YZ, and HC collected and grew the plants. HC and JH conducted the experiments. KW, BW, AE, JL, and YZ participated in the data analysis and manuscript preparation.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Responsible Editor: Andreas Houben
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Wang, K., Cheng, H., Han, J. et al. A comprehensive molecular cytogenetic analysis of the genome architecture in modern sugarcane cultivars. Chromosome Res 30, 29–41 (2022). https://doi.org/10.1007/s10577-021-09680-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-021-09680-3