Abstract
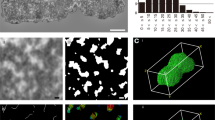
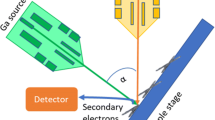
The chromosome compaction of chromatin fibers results in the formation of the nucleosome, which consists of a DNA unit coiled around a core of histone molecules associated with linker histone. The compaction of chromatin fibers has been a topic of controversy since the discovery of chromosomes in the 19th century. Although chromatin fibers were first identified using electron microscopy, the chromatin fibers on the surface of chromosome structures in plants remain unclear due to shrinking and breaking caused by prior chromosome isolation or preparation with alcohol and acid fixation, and critical point drying occurred into dehydration and denatured chromosomal proteins. This study aimed to develop a high-quality procedure for the isolation and preparation of plant chromosomes, maintaining the native chromosome structure, to elucidate the organization of chromatin fibers on the surface of plant chromosomes by electron microscopy. A simple technique to isolate intact barley (Hordeum vulgare) chromosomes with a high yield was developed, allowing chromosomes to be observed with a high-resolution scanning ion microscopy and helium ion microscopy (HIM) imaging technology, based on a scanning helium ion beam. HIM images from the surface chromatin fibers were analyzed to determine the size and alignment of the chromatin fibers. The unit size of the chromatin fibers was 11.6 ± 3.5 nm and was closely aligned to the chromatin network model. Our findings indicate that compacting the surface structure of barley via a chromatin network and observation via HIM are powerful tools for investigating the structure of chromatin.





Similar content being viewed by others
Data Availability
All data analyzed during this study are included in this published article.
Code availability
Not applicable.
Abbreviations
- APM:
-
Amiprophos methyl
- CPD:
-
Critical point drying
- EM:
-
Electron microscopy
- FIB/SEM:
-
Focused ion beam-SEM
- HIM:
-
Helium ion microscopy
- HU:
-
Hydroxyurea
- IL:
-
Ionic liquid
- PMSF:
-
Phenylmethylsulfonyl fluoride
- PI:
-
Propidium iodide
- SEM:
-
Scanning electron microscopy
- SHIM:
-
Scanning helium ion microscopy
- SEs:
-
Secondary electrons (SEs)
- TEM:
-
Transmission microscopy (TEM)
- DTT:
-
1,4-Dithiothreitol (DTT)
- DAPI:
-
4’,6-Diamidino-2-phenylindole (DAPI)
References
Adams RL, Lindsay JG (1967) Hydroxyurea reversal of inhibition and use as a cell-synchronizing agent. J Biol Chem 242:1314–1317
Baggott A, Mazaheri M, Zhou Y, Zhang H, Inkson BJ (2018) A comparison of He and Ne FIB imaging of cracks in microindented silicon nitride. Mater Charact 141:362–369. https://doi.org/10.1016/j.matchar.2018.05.006
Baldi S, Korber P, Becker PB (2020) Beads on a string—nucleosome array arrangements and folding of the chromatin fiber. Nat Struct Mol Biol 27:109–118. https://doi.org/10.1038/s41594-019-0368-x
Bandara CD, Ballerin G, Leppänen M, et al (2020) Resolving bio-nano interactions of E. coli bacteria-dragonfly wing interface with helium ion and 3D-structured illumination microscopy to understand bacterial death on nanotopography. ACS Biomater Sci Eng. https://doi.org/10.1021/acsbiomaterials.9b01973
Baxter J, Aragón L (2012) A model for chromosome condensation based on the interplay between condensin and topoisomerase II. Trends Genet 28:110–117. https://doi.org/10.1016/j.tig.2011.11.004
Bazou D, Behan G, Reid C et al (2011) Imaging of human colon cancer cells using He-Ion scanning microscopy. J Microsc 242:290–294. https://doi.org/10.1111/j.1365-2818.2010.03467.x
Bell DC (2009) Contrast mechanisms and image formation in helium ion microscopy. Microsc Microanal 15:147–153. https://doi.org/10.1017/s1431927609090138
Boopathi R, Dimitrov S, Hamiche A, Petosa C, Bednar J (2020) Cryo-electron microscopy of the chromatin fiber. Curr Opin Struct Biol 64:97–103. https://doi.org/10.1016/j.sbi.2020.06.016
Brock TD (1984) How sensitive is the light microscope for observations on microorganisms in natural habitats? Microb Ecol 10:297–300. https://doi.org/10.1007/bf02015555
Chamla Y (1988) C-anaphases in lymphocyte cultures versus premature centromere division syndromes. Hum Genet 78:111–114. https://doi.org/10.1007/bf00278177
Cleveland DW, Mao Y, Sullivan KF (2003) Centromeres and kinetochores: from epigenetics to mitotic checkpoint signaling. Cell 112:407–421. https://doi.org/10.1016/s0092-8674(03)00115-6
Clode PL (2006) Charge contrast imaging of biomaterials in a variable pressure scanning electron microscope. J Struct Biol 155:505–511. https://doi.org/10.1016/j.jsb.2006.04.004
Daban J-R (2015) Stacked thin layers of metaphase chromatin explain the geometry of chromosome rearrangements and banding. Sci Rep 5:14891. https://doi.org/10.1038/srep14891
De Laat AMM, Blaas J (1984) Flow-cytometric characterization and sorting of plant chromosomes. Theor Appl Genet 67:463–467. https://doi.org/10.1007/bf00263414
Doležel J, Binarová P, Lcretti S (1989) Analysis of nuclear DNA content in plant cells by flow cytometry. Biol Plant 31:113–120. https://doi.org/10.1007/bf02907241
Doležel J, Číhalíková J, Lucretti S (1992) A high-yield procedure for isolation of metaphase chromosomes from root tips of Vicia faba L. Planta 188:93–98. https://doi.org/10.1007/bf00198944
Dwiranti A, Lin L, Mochizuki E, Kuwabata S, Takaoka A, Uchiyama S, Fukui K (2012) Chromosome observation by scanning electron microscopy using ionic liquid. Microsc Res Tech 75:1113–1118. https://doi.org/10.1002/jemt.22038
Dwiranti A, Hamano T, Takata H, Nagano S, Guo H, Onishi K, Wako T, Uchiyama S, Fukui K (2014) The effect of magnesium ions on chromosome structure as observed by helium ion microscopy. Microsc Microanal 20:184–188. https://doi.org/10.1017/S1431927613013792
Gibcus JH, Samejima K, Goloborodko A, Samejima I, Naumova N, Nuebler J, Kanemaki MT, Xie L, Paulson JR, Earnshaw WC, Mirny LA, Dekker J (2018) A pathway for mitotic chromosome formation. Science 359:1–12. https://doi.org/10.1126/science.aao6135
Griesbach RJ, Malmberg RL, Carlson PS (1982) An improved technique for the isolation of higher plant chromosomes. Plant Sci Lett 24:55–60. https://doi.org/10.1016/0304-4211(82)90008-6
Hadlaczky G, Bisztray G, Praznovszky T, Dudits D (1983) Mass isolation of plant chromosomes and nuclei. Planta 157:278–285. https://doi.org/10.1007/BF00405195
Hamano T, Dwiranti A, Kaneyoshi K, Fukuda S, Kometani R, Nakao M, Takata H, Uchiyama S, Ohmido N, Fukui K (2014) Chromosome interior observation by focused ion beam/scanning electron microscopy (FIB/SEM) using ionic liquid technique. Microsc Microanal 20:1340–1347. https://doi.org/10.1017/s143192761401280x
Heu R, Shahbazmohamadi S, Yorston J, Capeder P (2019) Target material selection for sputter coating of SEM samples. MTO 27:32–36. https://doi.org/10.1017/s1551929519000610
Heyden HWv, Zachau HG (1971) Characterization of RNA in fractions of calf thymus chromatin. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis 232(4):651–660
Hill R, Rahman FF (2011) Advances in helium ion microscopy. Nucl Inst Methods Phys Res B 645:96–101. https://doi.org/10.1016/j.nima.2010.12.123
Hoagland DR, Arnon DI (1950) The water-culture method for growing plants without soil. Calif Univ Agric Exp Stn Circ Calif Agric Exp Stn Circ 347:1884–1949. https://doi.org/10.1007/s00412-007-0102-z
Houben A, Schroeder-Reiter E, Nagaki K, Nasuda S, Wanner G, Murata M, Endo TR (2007) CENH3 interacts with the centromeric retrotransposon cereba and GC-rich satellites and locates to centromeric substructures in barley. Chromosoma 116:275–283. https://doi.org/10.1007/s00412-007-0102-z
Huberman JA, Attardi G (1966) Isolation of metaphase chromosomes from HeLa cells. J Cell Biol 31(1):95–105
Iwano M, Fukui K, Takaichi S, Isogai A (1997) Globular and fibrous structure in barley chromosomes revealed by high-resolution scanning electron microscopy. Chromosome Sci 5:341–349. https://doi.org/10.1023/b:chro.0000038766.53836.c3
Joens MS, Huynh C, Kasuboski JM, Ferranti D, Sigal YJ, Zeitvogel F, Obst M, Burkhardt CJ, Curran KP, Chalasani SH, Stern LA, Goetze B, Fitzpatrick JAJ (2013) Helium ion microscopy (HIM) for the imaging of biological samples at sub-nanometer resolution. Sci Rep 3:3514. https://doi.org/10.1038/srep03514
Juarez T, Schroer A, Schwaiger R, Hodge AM (2018) Evaluating sputter deposited metal coatings on 3D printed polymer micro-truss structures. Mater Des 140:442–450. https://doi.org/10.1016/j.matdes.2017.12.005
Kaneyoshi K, Fukuda S, Dwiranti A et al (2015) Effects of dehydration and drying steps on human chromosome interior revealed by focused ion beam/scanning electron microscopy (FIB/SEM). Chromosome Sci 18:23–28. https://doi.org/10.11352/scr.18.23
Kixmoeller K, Allu PK, Black BE (2020) The centromere comes into focus: from CENP-A nucleosomes to kinetochore connections with the spindle. Open Biol 10:200051. https://doi.org/10.1098/rsob.200051
Legland D, Arganda-Carreras I, Andrey P (2016) MorphoLibJ: integrated library and plugins for mathematical morphology with ImageJ. Bioinformatics 32:3532–3534. https://doi.org/10.1093/bioinformatics/btw413
Lysák MA, ČíUhalíková J, Kubaláková M, Šimková H, Künzel G, Doležel J (1999) Flow karyotyping and sorting of mitotic chromosomes of barley (Hordeum vulgare L.). Chromosome Sci 7:431–444. https://doi.org/10.1023/a:1009293628638
Maeshima K, Imai R, Tamura S, Nozaki T (2014) Chromatin as dynamic 10-nm fibers. Chromosoma 123:225–237. https://doi.org/10.1007/s00412-014-0460-2
Manzanero S, Rutten T, Kotseruba V, Houben A (2002) Alterations in the distribution of histone H3 phosphorylation in mitotic plant chromosomes in response to cold treatment and the protein phosphatase inhibitor cantharidin. Chromosom Res 10:467–476. https://doi.org/10.1023/A:1020940313841
Martin R, Busch W, Herrmann RG, Wanner G (1994) Efficient preparation of plant chromosomes for high-resolution scanning electron microscopy. Chromosom Res 2:411–415. https://doi.org/10.1007/bf01552801
McGhee JD, Nickol JM, Felsenfeld G, Rau DC (1983) Higher order structure of chromatin: orientation of nucleosomes within the 30 nm chromatin solenoid is independent of species and spacer length. Cell 33:831–841. https://doi.org/10.1016/0092-8674(83)90025-9
Migneault I, Dartiguenave C, Bertrand MJ, Waldron KC (2004) Glutaraldehyde: behavior in aqueous solution, reaction with proteins, and application to enzyme crosslinking. Biotechniques 37:790–802. https://doi.org/10.1021/acs.jpcc.8b05383.s001
Mii M, Saxena PK, Fowke LC, King J (1987) Isolation of chromosomes from cell suspension cultures of Vicia hajastana. CYTOLOGIA 52:523–528. https://doi.org/10.1508/cytologia.52.523
Morejohn LC, Fosket DE (1984) Inhibition of plant microtubule polymerization in vitro by the phosphoric amide herbicide amiprophos-methyl. Science 224:874–876. https://doi.org/10.1126/science.224.4651.874
Morse RH, Simpson RT (1988) DNA in the nucleosome. Cell 54:285–287. https://doi.org/10.1016/0092-8674(88)90190-0
Ohmido N, Kijima K, Hoshi O, Ushiki T, Fukui K (2005) Comparison of surface structures between extended and condensed stages of barley chromosomes revealed with atomic force microscopy. CYTOLOGIA 70:101–108. https://doi.org/10.1508/cytologia.70.101
Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, O’Shea CC (2017) ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science 357:eaag0025. https://doi.org/10.1126/science.aag0025
Paulson JR, Laemmli UK (1977) The structure of histone-depleted metaphase chromosomes. Cell 12:817–828. https://doi.org/10.1016/0092-8674(77)90280-x
Poirier MG, Marko JF (2002) Mitotic chromosomes are chromatin networks without a mechanically contiguous protein scaffold. PNAS 99:15393–15397. https://doi.org/10.1073/PNAS.232442599
Pooley AS, Pardon JF, Richards BM (1974) The relation between the unit thread of chromosomes and isolated nucleohistone. J Mol Biol 85(4):533–549
Poonperm R, Takata H, Hamano T, Matsuda A, Uchiyama S, Hiraoka Y, Fukui K (2015) Chromosome scaffold is a double-stranded assembly of scaffold proteins. Sci Rep 5:1–10. https://doi.org/10.1038/srep11916
Sawafta F, Carlsen AT, Hall AR (2014) Membrane thickness dependence of nanopore formation with a focused helium ion beam. J Sens 14:8150–8161. https://doi.org/10.3390/s140508150
Schneider CA, Rasband WS, Eliceiri KW (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675. https://doi.org/10.1038/nmeth.2089
Schroeder-Reiter E (2004) High resolution analysis of mitotic metaphase chromosomes with scanning electron microscopy: localizing histone H3 modifications with immunogold labeling in barley (Hordeum vulgare). Verlag nicht ermittelbar
Schroeder-Reiter E, Pérez-Willard F, Zeile U, Wanner G (2009) Focused ion beam (FIB) combined with high resolution scanning electron microscopy: a promising tool for 3D analysis of chromosome architecture. J Struct Biol 165:97–106. https://doi.org/10.1016/j.jsb.2008.10.002
Schroeder-Reiter E, Sanei M, Houben A, Wanner G (2012) Current SEM techniques for de-and re-construction of centromeres to determine 3D CENH3 distribution in barley mitotic chromosomes. J Microsc 246:96–106. https://doi.org/10.1111/j.1365-2818.2011.03592.x
Sedat J, Manuelidis L (1978) A direct approach to the structure of eukaryotic chromosomes. In: Cold Spring Harbor symposia on quantitative biology. Cold Spring Harbor Laboratory Press, pp 331–350. https://doi.org/10.1101/sqb.1978.042.01.035
Segal E, Fondufe-Mittendorf Y, Chen L, Thåström AC, Field Y, Moore IK, Wang JPZ, Widom J (2006) A genomic code for nucleosome positioning. Nature 442:772–778. https://doi.org/10.1038/nature04979
Shih TY, Bonner J (1969) Chromosomal RNA of calf thymus chromatin. Biochimica et Biophysica Acta (BBA) - Nucleic Acids and Protein Synthesis 182(1):30–35
Skinner LG, Ockey CH (1971) Isolation, fractionation and biochemical analysis of the metaphase chromosomes of Microtus agrestis. Chromosoma 35(2):125–142
Squarzoni S, Cinti C, Santi S, Valmori A, Maraldi NM (1994) Preparation of chromosome spreads for electron (TEM, SEM, STEM), light and confocal microscopy. Chromosoma 103:381–392. https://doi.org/10.1007/bf00362282
Steger C (1998) An unbiased detector of curvilinear structures. IEEE PAMI 20:113–125. https://doi.org/10.1109/34.659930
Sugiyama S, Yoshino T, Kanahara H, Shichiri M, Fukushi D, Ohtani T (2004) Effects of acetic acid treatment on plant chromosome structures analyzed by atomic force microscopy. Anal Biochem 324:39–44. https://doi.org/10.1016/j.ab.2003.09.026
Swedlow JR, Hirano T (2003) The making of the mitotic chromosome: modern insights into classical questions. Component 11:557–569. https://doi.org/10.7554/elife.22280.023
Szabados L, Hadlaczky G, Dudits D (1981) Uptake of isolated plant chromosomes by plant protoplasts. Planta 151:141–145. https://doi.org/10.1007/bf00387815
Uchiyama S, Kobayashi S, Takata H et al (2005) Proteome analysis of human metaphase chromosomes. J Biol Chem 280:16994–17004. https://doi.org/10.1201/9781420044928-14
Wako T, Yoshida A, Kato J, Otsuka Y, Ogawa S, Kaneyoshi K, Takata H, Fukui K (2020) Human metaphase chromosome consists of randomly arranged chromatin fibres with up to 30-nm diameter. Sci Rep 10:8948. https://doi.org/10.1038/s41598-020-65842-z
Wanner G, Formanek H (2000) A new chromosome model. Nature 132:147–161. https://doi.org/10.1038/203343a0
Wanner G, Formanek H, Martin R, Herrmann RG (1991) High resolution scanning electron microscopy of plant chromosomes. Chromosoma 100:103–109. https://doi.org/10.1007/BF00418243
Wanner G, Schroeder-Reiter E, Formanek H (2005) 3D analysis of chromosome architecture: advantages and limitations with SEM. Cytogenet Genome Res 109:70–78. https://doi.org/10.1159/000082384
Wanner G, Schroeder-Reiter E, Ma W, Houben A, Schubert V (2015) The ultrastructure of mono-and holocentric plant centromeres: an immunological investigation by structured illumination microscopy and scanning electron microscopy. Chromosoma 124:503–517. https://doi.org/10.1007/s00412-015-0521-1
Williams S, Zhang X, Jacobsen C et al (1993) Measurements of wet metaphase chromosomes in the scanning transmission X-ray microscope. J Microsc 170:155–165. https://doi.org/10.1111/j.1365-2818.1993.tb03335.x
Woodcock CL, Ghosh RP (2010) Chromatin higher-order structure and dynamics. Cold Spring Harb Perspect Biol 2:a000596. https://doi.org/10.1101/cshperspect.a000596
Yih L-H, Lee T-C (2003) Induction of C-anaphase and diplochromosome through dysregulation of spindle assembly checkpoint by sodium arsenite in human fibroblasts. Cancer Res 63:6680–6688
Zhou Z, Li K, Yan R, Yu G, Gilpin CJ, Jiang W, Irudayaraj JMK (2019) The transition structure of chromatin fibers at the nanoscale probed by cryogenic electron tomography. Nanoscale 11:13783–13789. https://doi.org/10.1039/c9nr02042j
Acknowledgements
We would like to thank Dr. Kimihiro Norizawa, the Nanotechnology Open Facilities Center in Osaka University for all his guidance regarding the HIM technique. We also gratefully acknowledge scholarship support provided by the Japanese Ministry of Education, Culture, Sports, Science, and Technology (to CS). We would like to thank Editage (www.editage.com) for English language editing.
Funding
This work was supported by “e-ASIA Joint Research Program,” “Development of nanovisualization for structural analyses of genetic materials and early infection process for further innovation of functional bio-nanotechnology” from SICORP and the Mobility Plus Project (grant no. JSPS-18-07) (to NO). This work was also supported by “The 51th Research Grants in the Natural Sciences” from The Mitsubishi Foundation (to NO) and Nanotechnology Platform of MEXT, Grant Number JPMXP09F20OS0004 (to NO).
Author information
Authors and Affiliations
Contributions
NO and KF conceived and designed research. CS and RP conducted experiments. TW contributed the development of image analysis tools. CS analyzed data. CS and NO wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
All authors listed had contributed sufficiently to the project to be included as authors, and all those who are qualified to be authors are listed in the author byline. We declare no competing interests among all authors and project sources.
Additional information
Responsible Editor: Beth Sullivan
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Sartsanga, C., Phengchat, R., Fukui, K. et al. Surface structures consisting of chromatin fibers in isolated barley (Hordeum vulgare) chromosomes revealed by helium ion microscopy. Chromosome Res 29, 81–94 (2021). https://doi.org/10.1007/s10577-021-09649-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-021-09649-2