Abstract
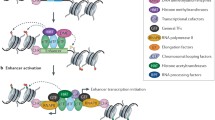
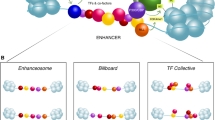
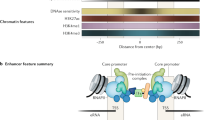
Enhancers are traditionally viewed as DNA sequences located some distance from a promoter that act in cis and in an orientation-independent fashion to increase utilization of specific promoters and thereby regulate gene expression. Much progress has been made over the last decade toward understanding how these distant elements interact with target promoters, but how transcription is enhanced remains an object of active inquiry. Recent reports convey the prevalence and diversity of enhancer transcription and transcripts and support both as key factors with mechanistically distinct, but not mutually exclusive roles in enhancer function. Decoupling the causes and effects of transcription on the local chromatin landscape and understanding the role of enhancer transcripts in the context of long-range interactions are challenges that require additional attention. In this review, we focus on the possible functions of enhancer transcription by highlighting several recent enhancer RNA papers and, within the context of other enhancer studies, speculate on the role of enhancer transcription in regulating differential gene expression.





Similar content being viewed by others
Abbreviations
- AR:
-
Androgen receptor
- Arc:
-
Activity-regulated cytoskeleton-associated protein
- CBP:
-
CREB-binding protein
- ChIP:
-
Chromatin immunoprecipitation
- ChIP-seq:
-
ChIP coupled with massively paralleled sequencing
- E2:
-
17β-Estradiol
- ER-α:
-
Estrogen receptor-α
- eRNA:
-
Enhancer RNA
- GRO-seq:
-
Global run-on sequencing
- H3K9ac:
-
Histone H3 acetylated at lysine 9
- H3K27ac:
-
Histone H3 acetylated at lysine 27
- H3K4me1:
-
Histone H3 monomethylated at lysine 4
- H3K4me2:
-
Histone H3 dimethylated at lysine 4
- H3K4me3:
-
Histone H3 trimethylated at lysine 4
- H3K36me3:
-
Histone H3 trimethylated at lysine 36
- H3S10ph:
-
H3 serine-10 phosphorylation
- HAT:
-
Histone acetyltransferase
- HDAC:
-
Histone deacetylase
- kb:
-
Kilobase
- KCl:
-
Potassium chloride
- KLA:
-
Kdo2-lipid A
- LNA:
-
Locked nucleic acids
- lncRNA:
-
Long noncoding RNA
- LPS:
-
Lipopolysaccharide
- ncRNA-a:
-
Noncoding RNA-activating
- NPAS4:
-
Neuronal PAS domain 4
- Pol II:
-
RNA polymerase II
- RNA-seq:
-
Massively parallel RNA sequencing
- SRF:
-
Serum response factor
- siRNA:
-
Small interfering RNA
- TF:
-
Transcription factor
- TLR4:
-
Toll-like receptor 4
- TSS:
-
Transcription start sites
- TTX:
-
Tetrodotoxin
- 3C:
-
Chromosome conformation capture
- 3D-DSL:
-
Three-dimensional DNA selection and ligation
- 4C:
-
Circularized chromosome conformation capture
References
Barski A, Cuddapah S, Cui K et al (2007) High-resolution profiling of histone methylations in the human genome. Cell 129:823–837
Bonn S, Zinzen RP, Girardot C et al (2012) Tissue-specific analysis of chromatin state identifies temporal signatures of enhancer activity during embryonic development. Nat Genet 44:148–156
Calo E, Wysocka J (2013) Modification of enhancer chromatin: what, how, and why? Mol Cell 49:825–837
Consortium EP, Birney E, Stamatoyannopoulos JA et al (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature 447:799–816
Core LJ, Waterfall JJ, Lis JT (2008) Nascent RNA sequencing reveals widespread pausing and divergent initiation at human promoters. Science 322:1845–1848
Creyghton MP, Cheng AW, Welstead GG et al (2010) Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc Natl Acad Sci U S A 107:21931–21936
De Santa F, Barozzi I, Mietton F et al (2010) A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol 8:e1000384
Djebali S, Davis CA, Merkel A et al (2012) Landscape of transcription in human cells. Nature 489:101–108
Ernst P, Wang J, Huang M, Goodman RH, Korsmeyer SJ (2001) MLL and CREB bind cooperatively to the nuclear coactivator CREB-binding protein. Mol Cell Biol 21:2249–2258
Ernst J, Kheradpour P, Mikkelsen TS et al (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473:43–49
Fullwood MJ, Liu MH, Pan YF et al (2009) An oestrogen-receptor-alpha-bound human chromatin interactome. Nature 462:58–64
Goto NK, Zor T, Martinez-Yamout M, Dyson HJ, Wright PE (2002) Cooperativity in transcription factor binding to the coactivator CREB-binding protein (CBP)—the mixed lineage leukemia protein (MLL) activation domain binds to an allosteric site on the KIX domain. J Biol Chem 277:43168–43174
Guttman M, Amit I, Garber M et al (2009) Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature 458:223–227
Hah N, Murakami S, Nagari A, Danko CG, Kraus WL (2013) Enhancer transcripts mark active estrogen receptor binding sites. Genome Res 23:1210–1223
He HH, Meyer CA, Shin H et al (2010) Nucleosome dynamics define transcriptional enhancers. Nat Genet 42:343–347
Heintzman ND, Stuart RK, Hon G et al (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat Genet 39:311–318
Heintzman ND, Hon GC, Hawkins RD et al (2009) Histone modifications at human enhancers reflect global cell-type-specific gene expression. Nature 459:108–112
Heinz S, Benner C, Spann N et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589
Ho Y, Elefant F, Liebhaber SA, Cooke NE (2006) Locus control region transcription plays an active role in long-range gene activation. Mol Cell 23:365–375
Hwang YC, Zheng Q, Gregory BD, Wang LS (2013) High-throughput identification of long-range regulatory elements and their target promoters in the human genome. Nucleic Acids Res 41:4835–4846
John S, Sabo PJ, Thurman RE et al (2011) Chromatin accessibility pre-determines glucocorticoid receptor binding patterns. Nat Genet 43:264–U116
Kagey MH, Newman JJ, Bilodeau S et al (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467:430–435
Kaikkonen MU, Spann NJ, Heinz S et al (2013) Remodeling of the enhancer landscape during macrophage activation is coupled to enhancer transcription. Mol Cell 51:310–325
Kim A, Zhao H, Ifrim I, Dean A (2007) Beta-globin intergenic transcription and histone acetylation dependent on an enhancer. Mol Cell Biol 27:2980–2986
Kim TK, Hemberg M, Gray JM et al (2010) Widespread transcription at neuronal activity-regulated enhancers. Nature 465:182–187
Koch CM, Andrews RM, Flicek P et al (2007) The landscape of histone modifications across 1% of the human genome in five human cell lines. Genome Res 17:691–707
Koch F, Fenouil R, Gut M et al (2011) Transcription initiation platforms and GTF recruitment at tissue-specific enhancers and promoters. Nat Struct Mol Biol 18:956–963
Kornienko AE, Guenzl PM, Barlow DP, Pauler FM (2013) Gene regulation by the act of long non-coding RNA transcription. BMC Biol 11:59
Kowalczyk MS, Hughes JR, Garrick D et al (2012) Intragenic enhancers act as alternative promoters. Mol Cell 45:447–458
Lai F, Orom UA, Cesaroni M et al (2013) Activating RNAs associate with Mediator to enhance chromatin architecture and transcription. Nature 494:497–501
Lam MT, Cho H, Lesch HP et al (2013) Rev-Erbs repress macrophage gene expression by inhibiting enhancer-directed transcription. Nature 498:511–515
Li G, Ruan X, Auerbach RK et al (2012) Extensive promoter-centered chromatin interactions provide a topological basis for transcription regulation. Cell 148:84–98
Li W, Notani D, Ma Q et al (2013) Functional roles of enhancer RNAs for oestrogen-dependent transcriptional activation. Nature 498:516–520
Lupien M, Eeckhoute J, Meyer CA et al (2008) FoxA1 translates epigenetic signatures into enhancer-driven lineage-specific transcription. Cell 132:958–970
Melo CA, Drost J, Wijchers PJ et al (2013) eRNAs are required for p53-dependent enhancer activity and gene transcription. Mol Cell 49:524–535
Mikkelsen TS, Ku MC, Jaffe DB et al (2007) Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature 448:553–U552
Orom UA, Derrien T, Beringer M et al (2010) Long noncoding RNAs with enhancer-like function in human cells. Cell 143:46–58
Ostuni R, Piccolo V, Barozzi I et al (2013) Latent enhancers activated by stimulation in differentiated cells. Cell 152:157–171
Petesch SJ, Lis JT (2012) Overcoming the nucleosome barrier during transcript elongation. Trends Genet 28:285–294
Rada-Iglesias A, Bajpai R, Swigut T et al (2011) A unique chromatin signature uncovers early developmental enhancers in humans. Nature 470:279–283
Sandhu KS, Li GL, Poh HM et al (2012) Large-scale functional organization of long-range chromatin interaction networks. Cell Rep 2:1207–1219
Sanyal A, Lajoie BR, Jain G, Dekker J (2012) The long-range interaction landscape of gene promoters. Nature 489:109–113
Schmidt D, Schwalie PC, Ross-Innes CS et al (2010) A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res 20:578–588
Thurman RE, Rynes E, Humbert R et al (2012) The accessible chromatin landscape of the human genome. Nature 489:75–82
Tsai MC, Manor O, Wan Y et al (2010) Long noncoding RNA as modular scaffold of histone modification complexes. Science 329:689–693
Tuan D, Kong SM, Hu K (1992) Transcription of the hypersensitive site Hs2 enhancer in erythroid-cells. Proc Natl Acad Sci U S A 89:11219–11223
Visel A, Blow MJ, Li Z et al (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457:854–858
Wang Q, Carroll JS, Brown M (2005) Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol Cell 19:631–642
Wang Z, Zang C, Rosenfeld JA et al (2008) Combinatorial patterns of histone acetylations and methylations in the human genome. Nat Genet 40:897–903
Wang D, Garcia-Bassets I, Benner C et al (2011) Reprogramming transcription by distinct classes of enhancers functionally defined by eRNA. Nature 474:390-+
Wang J, Zhuang JL, Iyer S et al (2012) Sequence features and chromatin structure around the genomic regions bound by 119 human transcription factors. Genome Res 22:1798–1812
Zaret KS, Carroll JS (2011) Pioneer transcription factors: establishing competence for gene expression. Genes Dev 25:2227–2241
Zentner GE, Tesar PJ, Scacheri PC (2011) Epigenetic signatures distinguish multiple classes of enhancers with distinct cellular functions. Genome Res 21:1273–1283
Acknowledgments
The authors apologize to those whose work is not cited due to space limitations. This work was supported by the National Institutes of Health [GM073120 and NS080779 to B.P.C].
Conflict of interest
The authors declare they have no conflicts of interest.
Author information
Authors and Affiliations
Corresponding author
Additional information
Responsible Editor: Brian P. Chadwick, Kristin C. Scott, and Beth A. Sullivan
Rights and permissions
About this article
Cite this article
Darrow, E.M., Chadwick, B.P. Boosting transcription by transcription: enhancer-associated transcripts. Chromosome Res 21, 713–724 (2013). https://doi.org/10.1007/s10577-013-9384-6
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10577-013-9384-6