Abstract
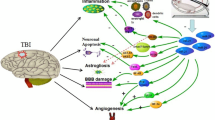
History of traumatic brain injury (TBI) represents a significant risk factor for development of dementia and neurodegenerative disorders in later life. While histopathological sequelae and neurological diagnostics of TBI are well defined, the molecular events linking the post-TBI signaling and neurodegenerative cascades remain unknown. It is not only due to the brain’s inaccessibility to direct molecular analysis but also due to the lack of well-defined and highly informative peripheral biomarkers. MicroRNAs (miRNAs) in blood are promising candidates to address this gap. Using integrative bioinformatics pipeline including miRNA:target identification, pathway enrichment, and protein–protein interactions analysis we identified set of genes, interacting proteins, and pathways that are connected to previously reported peripheral miRNAs, deregulated following severe traumatic brain injury (sTBI) in humans. This meta-analysis revealed a spectrum of genes closely related to critical biological processes, such as neuroregeneration including axon guidance and neurite outgrowth, neurotransmission, inflammation, proliferation, apoptosis, cell adhesion, and response to DNA damage. More importantly, we have identified molecular pathways associated with neurodegenerative conditions, including Alzheimer’s and Parkinson’s diseases, based on purely peripheral markers. The pathway signature after acute sTBI is similar to the one observed in chronic neurodegenerative conditions, which implicates a link between the post-sTBI signaling and neurodegeneration. Identified key hub interacting proteins represent a group of novel candidates for potential therapeutic targets or biomarkers.




Similar content being viewed by others
Abbreviations
- sTBI:
-
Severe traumatic brain injury
- AD:
-
Alzheimer’s disease
- miRNA:
-
MicroRNA
References
Alosco ML, Koerte IK, Tripodis Y, Mariani M, Chua AS, Jarnagin J, Rahimpour Y, Puzo C, Healy RC, Martin B, Chaisson CE, Cantu RC, Au R, McClean M, McKee AC, Lin AP, Shenton ME, Killiany RJ, Stern RA (2018) White matter signal abnormalities in former National Football League players. Alzheimers Dement (amst) 10:56–65. https://doi.org/10.1016/j.dadm.2017.10.003
Atif H, Hicks SD (2019) A review of microRNA biomarkers in traumatic brain injury. J Exp Neurosci 13:1179069519832286. https://doi.org/10.1177/1179069519832286
Barbagallo C, Mostile G, Baglieri G, Giunta F, Luca A, Raciti L, Zappia M, Purrello M, Ragusa M, Nicoletti A (2020) Specific signatures of serum mirnas as potential biomarkers to discriminate clinically similar neurodegenerative and vascular-related diseases. Cell Mol Neurobiol 40:531–546. https://doi.org/10.1007/s10571-019-00751-y
Bartolotti N, Bennett DA, Lazarov O (2016) Reduced pCREB in Alzheimer’s disease prefrontal cortex is reflected in peripheral blood mononuclear cells. Mol Psychiatry 21:1158–1166. https://doi.org/10.1038/mp.2016.111
van den Berg MMJ, Krauskopf J, Ramaekers JG, Kleinjans JCS, Prickaerts J, Briede JJ (2020) Circulating microRNAs as potential biomarkers for psychiatric and neurodegenerative disorders. Prog Neurobiol 185:101732. https://doi.org/10.1016/j.pneurobio.2019.101732
Bhomia M, Balakathiresan NS, Wang KK, Papa L, Maheshwari RK (2016) A panel of serum MiRNA biomarkers for the diagnosis of severe to mild traumatic brain injury in humans. Sci Rep 6:28148. https://doi.org/10.1038/srep28148
Blennow K, Brody DL, Kochanek PM, Levin H, McKee A, Ribbers GM, Yaffe K, Zetterberg H (2016) Traumatic brain injuries. Nat Rev Dis Primers 2:16084. https://doi.org/10.1038/nrdp.2016.84
Brown KR, Otasek D, Ali M, McGuffin MJ, Xie W, Devani B, Toch IL, Jurisica I (2009) NAViGaTOR: network analysis, visualization and graphing toronto. Bioinformatics 25:3327–3329. https://doi.org/10.1093/bioinformatics/btp595
Bryan MR, Bowman AB (2017) Manganese and the insulin-IGF signaling network in Huntington’s disease and other neurodegenerative disorders. Adv Neurobiol 18:113–142. https://doi.org/10.1007/978-3-319-60189-2_6
Chandran R, Sharma A, Bhomia M, Balakathiresan NS, Knollmann-Ritschel BE, Maheshwari RK (2017) Differential expression of microRNAs in the brains of mice subjected to increasing grade of mild traumatic brain injury. Brain Inj 31:106–119. https://doi.org/10.1080/02699052.2016.1213420
Chang CY, Liang MZ, Wu CC, Huang PY, Chen HI, Yet SF, Tsai JW, Kao CF, Chen L (2020) WNT3A promotes neuronal regeneration upon traumatic brain injury. Int J Mol Sci 21:1463. https://doi.org/10.3390/ijms21041463
Cheng L, Doecke JD, Sharples RA, Villemagne VL, Fowler CJ, Rembach A, Martins RN, Rowe CC, Macaulay SL, Masters CL, Hill AF, Australian Imaging B, Lifestyle Research G (2015) Prognostic serum miRNA biomarkers associated with Alzheimer’s disease shows concordance with neuropsychological and neuroimaging assessment. Mol Psychiatry 20:1188–1196. https://doi.org/10.1038/mp.2014.127
Chiaretti A, Antonelli A, Genovese O, Pezzotti P, Rocco CD, Viola L, Riccardi R (2008) Nerve growth factor and doublecortin expression correlates with improved outcome in children with severe traumatic brain injury. J Trauma 65:80–85. https://doi.org/10.1097/TA.0b013e31805f7036
Chio CC, Chang CP, Lin MT, Su FC, Yang CZ, Tseng HY, Liu ZM, Huang HS (2014) Involvement of TG-interacting factor in microglial activation during experimental traumatic brain injury. J Neurochem 131:816–824. https://doi.org/10.1111/jnc.12971
Cogswell JP, Ward J, Taylor IA, Waters M, Shi Y, Cannon B, Kelnar K, Kemppainen J, Brown D, Chen C, Prinjha RK, Richardson JC, Saunders AM, Roses AD, Richards CA (2008) Identification of miRNA changes in Alzheimer’s disease brain and CSF yields putative biomarkers and insights into disease pathways. J Alzheimers Dis 14:27–41. https://doi.org/10.3233/jad-2008-14103
Corps KN, Roth TL, McGavern DB (2015) Inflammation and neuroprotection in traumatic brain injury. JAMA Neurol 72:355–362. https://doi.org/10.1001/jamaneurol.2014.3558
Delic V, Beck KD, Pang KCH, Citron BA (2020) Biological links between traumatic brain injury and Parkinson’s disease. Acta Neuropathol Commun 8:45. https://doi.org/10.1186/s40478-020-00924-7
Denk J, Boelmans K, Siegismund C, Lassner D, Arlt S, Jahn H (2015) MicroRNA profiling of CSF reveals potential biomarkers to detect Alzheimer’s disease. PLoS ONE 10:e0126423. https://doi.org/10.1371/journal.pone.0126423
Dewan MC, Rattani A, Gupta S, Baticulon RE, Hung YC, Punchak M, Agrawal A, Adeleye AO, Shrime MG, Rubiano AM, Rosenfeld JV, Park KB (2019) Estimating the global incidence of traumatic brain injury. J Neurosurg 130:1080–1097. https://doi.org/10.3171/2017.10.JNS17352
Di Pietro V, Ragusa M, Davies D, Su Z, Hazeldine J, Lazzarino G, Hill LJ, Crombie N, Foster M, Purrello M, Logan A, Belli A (2017) MicroRNAs as novel biomarkers for the diagnosis and prognosis of mild and severe traumatic brain injury. J Neurotrauma 34:1948–1956. https://doi.org/10.1089/neu.2016.4857
Di Pietro V, Yakoub KM, Scarpa U, Di Pietro C, Belli A (2018) MicroRNA signature of traumatic brain injury: from the biomarker discovery to the point-of-care. Front Neurol 9:429. https://doi.org/10.3389/fneur.2018.00429
Edwards KA, Motamedi V, Osier ND, Kim HS, Yun S, Cho YE, Lai C, Dell KC, Carr W, Walker P, Ahlers S, LoPresti M, Yarnell A, Tschiffley A, Gill JM (2020) A Moderate blast exposure results in dysregulated gene network activity related to cell death, survival, structure, and metabolism. Front Neurol 11:91. https://doi.org/10.3389/fneur.2020.00091
Farr SA, Niehoff ML, Kumar VB, Roby DA, Morley JE (2019) Inhibition of glycogen synthase kinase 3beta as a treatment for the prevention of cognitive deficits after a traumatic brain injury. J Neurotrauma 36:1869–1875. https://doi.org/10.1089/neu.2018.5999
Forlenza OV, Diniz BS, Talib LL, Mendonca VA, Ojopi EB, Gattaz WF, Teixeira AL (2009) Increased serum IL-1beta level in Alzheimer’s disease and mild cognitive impairment. Dement Geriatr Cogn Disord 28:507–512. https://doi.org/10.1159/000255051
Gao W, Li F, Zhou Z, Xu X, Wu Y, Zhou S, Yin D, Sun D, Xiong J, Jiang R, Zhang J (2017) IL-2/Anti-IL-2 complex attenuates inflammation and BBB disruption in mice subjected to traumatic Brain injury. Front Neurol 8:281. https://doi.org/10.3389/fneur.2017.00281
Gardner RC, Burke JF, Nettiksimmons J, Goldman S, Tanner CM, Yaffe K (2015) Traumatic brain injury in later life increases risk for Parkinson disease. Ann Neurol 77:987–995. https://doi.org/10.1002/ana.24396
Gardner RC, Byers AL, Barnes DE, Li Y, Boscardin J, Yaffe K (2018) Mild TBI and risk of Parkinson disease: a chronic effects of neurotrauma consortium study. Neurology 90:e1771–e1779. https://doi.org/10.1212/WNL.0000000000005522
Golz C, Kirchhoff FP, Westerhorstmann J, Schmidt M, Hirnet T, Rune GM, Bender RA, Schafer MKE (2019) Sex hormones modulate pathogenic processes in experimental traumatic brain injury. J Neurochem 150:173–187. https://doi.org/10.1111/jnc.14678
Gu Q, Cuevas E, Raymick J, Kanungo J, Sarkar S (2020) Downregulation of 14–3-3 Proteins in Alzheimer’s disease. Mol Neurobiol 57:32–40. https://doi.org/10.1007/s12035-019-01754-y
Guo Z, Cupples LA, Kurz A, Auerbach SH, Volicer L, Chui H, Green RC, Sadovnick AD, Duara R, DeCarli C, Johnson K, Go RC, Growdon JH, Haines JL, Kukull WA, Farrer LA (2000) Head injury and the risk of AD in the MIRAGE study. Neurology 54:1316–1323. https://doi.org/10.1212/wnl.54.6.1316
Guo R, Fan G, Zhang J, Wu C, Du Y, Ye H, Li Z, Wang L, Zhang Z, Zhang L, Zhao Y, Lu Z (2017) A 9-microRNA signature in serum serves as a noninvasive biomarker in early diagnosis of Alzheime’’s disease. J Alzheimers Dis 60:1365–1377. https://doi.org/10.3233/JAD-170343
He XY, Dan QQ, Wang F, Li YK, Fu SJ, Zhao N, Wang TH (2018) Protein network analysis of the serum and their functional implication in patients subjected to traumatic brain injury. Front Neurosci 12:1049. https://doi.org/10.3389/fnins.2018.01049
Hu Z, Yu D, Almeida-Suhett C, Tu K, Marini AM, Eiden L, Braga MF, Zhu J, Li Z (2012) Expression of miRNAs and their cooperative regulation of the pathophysiology in traumatic brain injury. PLoS ONE 7:e39357. https://doi.org/10.1371/journal.pone.0039357
Huang CH, Lin CW, Lee YC, Huang CY, Huang RY, Tai YC, Wang KW, Yang SN, Sun YT, Wang HK (2018a) Is traumatic brain injury a risk factor for neurodegeneration? A meta-analysis of population-based studies. BMC Neurol 18:184. https://doi.org/10.1186/s12883-018-1187-0
Huang S, Ge X, Yu J, Han Z, Yin Z, Li Y, Chen F, Wang H, Zhang J, Lei P (2018b) Increased miR-124-3p in microglial exosomes following traumatic brain injury inhibits neuronal inflammation and contributes to neurite outgrowth via their transfer into neurons. FASEB J 32:512–528. https://doi.org/10.1096/fj.201700673R
Iankova A (2006) The Glasgow Coma Scale: clinical application in emergency departments. Emerg Nurse 14:30–35. https://doi.org/10.7748/en2006.12.14.8.30.c4221
Inestrosa NC, Varela-Nallar L (2014) Wnt signaling in the nervous system and in Alzheimer’s disease. J Mol Cell Biol 6:64–74. https://doi.org/10.1093/jmcb/mjt051
Johnson VE, Stewart W, Smith DH (2012) Widespread tau and amyloid-beta pathology many years after a single traumatic brain injury in humans. Brain Pathol 22:142–149. https://doi.org/10.1111/j.1750-3639.2011.00513.x
Johnson D, Cartagena CM, Tortella FC, Dave JR, Schmid KE, Boutte AM (2017) Acute and subacute microRNA dysregulation is associated with cytokine responses in the rodent model of penetrating ballistic-like brain injury. J Trauma Acute Care Surg 83:S145–S149. https://doi.org/10.1097/TA.0000000000001475
Koerte IK, Nichols E, Tripodis Y, Schultz V, Lehner S, Igbinoba R, Chuang AZ, Mayinger M, Klier EM, Muehlmann M, Kaufmann D, Lepage C, Heinen F, Schulte-Korne G, Zafonte R, Shenton ME, Sereno AB (2017) Impaired cognitive performance in youth athletes exposed to repetitive head impacts. J Neurotrauma 34:2389–2395. https://doi.org/10.1089/neu.2016.4960
Kotlyar M, Pastrello C, Ahmed Z, Chee J, Varyova Z, Jurisica I (2022) IID 2021: towards context-specific protein interaction analyses by increased coverage, enhanced annotation and enrichment analysis. Nucleic Acids Res 50:D640–D647. https://doi.org/10.1093/nar/gkab1034
Lai Y, Stange C, Wisniewski SR, Adelson PD, Janesko-Feldman KL, Brown DS, Kochanek PM, Clark RS (2006) Mitochondrial heat shock protein 60 is increased in cerebrospinal fluid following pediatric traumatic brain injury. Dev Neurosci 28:336–341. https://doi.org/10.1159/000094159
Letiembre M, Liu Y, Walter S, Hao W, Pfander T, Wrede A, Schulz-Schaeffer W, Fassbender K (2009) Screening of innate immune receptors in neurodegenerative diseases: a similar pattern. Neurobiol Aging 30:759–768. https://doi.org/10.1016/j.neurobiolaging.2007.08.018
Lewandowski SA, Fredriksson L, Lawrence DA, Eriksson U (2016) Pharmacological targeting of the PDGF-CC signaling pathway for blood-brain barrier restoration in neurological disorders. Pharmacol Ther 167:108–119. https://doi.org/10.1016/j.pharmthera.2016.07.016
Liu DZ, Sharp FR, Van KC, Ander BP, Ghiasvand R, Zhan X, Stamova B, Jickling GC, Lyeth BG (2014) Inhibition of SRC family kinases protects hippocampal neurons and improves cognitive function after traumatic brain injury. J Neurotrauma 31:1268–1276. https://doi.org/10.1089/neu.2013.3250
Lugli G, Cohen AM, Bennett DA, Shah RC, Fields CJ, Hernandez AG, Smalheiser NR (2015) Plasma exosomal miRNAs in persons with and without Alzheimer disease: altered expression and prospects for biomarkers. PLoS ONE 10:e0139233. https://doi.org/10.1371/journal.pone.0139233
Lusardi TA, Phillips JI, Wiedrick JT, Harrington CA, Lind B, Lapidus JA, Quinn JF, Saugstad JA (2017) MicroRNAs in human cerebrospinal fluid as biomarkers for Alzheimer’s disease. J Alzheimers Dis 55:1223–1233. https://doi.org/10.3233/JAD-160835
Ma J, Jiang Q, Xu J, Sun Q, Qiao Y, Chen W, Wu Y, Wang Y, Xiao Q, Liu J, Tang H, Chen S (2015) Plasma insulin-like growth factor 1 is associated with cognitive impairment in Parkinson’s disease. Dement Geriatr Cogn Disord 39:251–256. https://doi.org/10.1159/000371510
Manley G, Gardner AJ, Schneider KJ, Guskiewicz KM, Bailes J, Cantu RC, Castellani RJ, Turner M, Jordan BD, Randolph C, Dvorak J, Hayden KA, Tator CH, McCrory P, Iverson GL (2017) A systematic review of potential long-term effects of sport-related concussion. Br J Sports Med 51:969–977. https://doi.org/10.1136/bjsports-2017-097791
Matyasova K, Csicsatkova N, Filipcik P, Jurisica I, Cente M (2021) Peripheral microRNA alteration and pathway signaling after mild traumatic brain injury. Gen Physiol Biophys 40:523–539. https://doi.org/10.4149/gpb_2021038
McKee AC, Cantu RC, Nowinski CJ, Hedley-Whyte ET, Gavett BE, Budson AE, Santini VE, Lee HS, Kubilus CA, Stern RA (2009) Chronic traumatic encephalopathy in athletes: progressive tauopathy after repetitive head injury. J Neuropathol Exp Neurol 68:709–735. https://doi.org/10.1097/NEN.0b013e3181a9d503
McKee AC, Abdolmohammadi B, Stein TD (2018) The neuropathology of chronic traumatic encephalopathy. Handb Clin Neurol 158:297–307. https://doi.org/10.1016/B978-0-444-63954-7.00028-8
Menon DK, Schwab K, Wright DW, Maas AI, Demographics and Clinical Assessment Working Group of the International Interagency Initiative toward Common Data Elements for Research on Traumatic Brain Injury and Psychological Health (2010) Position statement: definition of traumatic brain injury. Arch Phys Med Rehabil 91:1637–1640. https://doi.org/10.1016/j.apmr.2010.05.017
Merlo L, Cimino F, Angileri FF, La Torre D, Conti A, Cardali SM, Saija A, Germano A (2014) Alteration in synaptic junction proteins following traumatic brain injury. J Neurotrauma 31:1375–1385. https://doi.org/10.1089/neu.2014.3385
Miranda M, Morici JF, Zanoni MB, Bekinschtein P (2019) Brain-derived neurotrophic factor: a key molecule for memory in the healthy and the pathological brain. Front Cell Neurosci 13:363. https://doi.org/10.3389/fncel.2019.00363
Mohamed AZ, Nestor PJ, Cumming P, Nasrallah FA, Alzheimer’s Disease Neuroimaging I (2022) Traumatic brain injury fast-forwards Alzheimer’s pathology: evidence from amyloid positron emission tomorgraphy imaging. J Neurol 269:873–884. https://doi.org/10.1007/s00415-021-10669-5
Nagaraj S, Laskowska-Kaszub K, Debski KJ, Wojsiat J, Dabrowski M, Gabryelewicz T, Kuznicki J, Wojda U (2017) Profile of 6 microRNA in blood plasma distinguish early stage Alzheimer’s disease patients from non-demented subjects. Oncotarget 8:16122–16143. https://doi.org/10.18632/oncotarget.15109
Nordstrom P, Michaelsson K, Gustafson Y, Nordstrom A (2014) Traumatic brain injury and young onset dementia: a nationwide cohort study. Ann Neurol 75:374–381. https://doi.org/10.1002/ana.24101
Norsworthy PJ, Thompson AGB, Mok TH, Guntoro F, Dabin LC, Nihat A, Paterson RW, Schott JM, Collinge J, Mead S, Vire EA (2020) A blood miRNA signature associates with sporadic Creutzfeldt-Jakob disease diagnosis. Nat Commun 11:3960. https://doi.org/10.1038/s41467-020-17655-x
O’Connell GC, Smothers CG, Winkelman C (2020) Bioinformatic analysis of brain-specific miRNAs for identification of candidate traumatic brain injury blood biomarkers. Brain Inj 34:965–974. https://doi.org/10.1080/02699052.2020.1764102
Oliva AA Jr, Kang Y, Sanchez-Molano J, Furones C, Atkins CM (2012) STAT3 signaling after traumatic brain injury. J Neurochem 120:710–720. https://doi.org/10.1111/j.1471-4159.2011.07610.x
Omalu BI, DeKosky ST, Minster RL, Kamboh MI, Hamilton RL, Wecht CH (2005) Chronic traumatic encephalopathy in a National Football League player. Neurosurgery 57:128–134. https://doi.org/10.1227/01.neu.0000163407.92769.ed
Pierce JE, Trojanowski JQ, Graham DI, Smith DH, McIntosh TK (1996) Immunohistochemical characterization of alterations in the distribution of amyloid precursor proteins and beta-amyloid peptide after experimental brain injury in the rat. J Neurosci 16:1083–1090. https://doi.org/10.1523/JNEUROSCI.16-03-01083.1996
Pinero J, Ramirez-Anguita JM, Sauch-Pitarch J, Ronzano F, Centeno E, Sanz F, Furlong LI (2020) The DisGeNET knowledge platform for disease genomics: 2019 update. Nucleic Acids Res 48:D845–D855. https://doi.org/10.1093/nar/gkz1021
Plassman BL, Havlik RJ, Steffens DC, Helms MJ, Newman TN, Drosdick D, Phillips C, Gau BA, Welsh-Bohmer KA, Burke JR, Guralnik JM, Breitner JC (2000) Documented head injury in early adulthood and risk of Alzheimer’s disease and other dementias. Neurology 55:1158–1166. https://doi.org/10.1212/wnl.55.8.1158
Qin X, Li L, Lv Q, Shu Q, Zhang Y, Wang Y (2018) Expression profile of plasma microRNAs and their roles in diagnosis of mild to severe traumatic brain injury. PLoS ONE 13:e0204051. https://doi.org/10.1371/journal.pone.0204051
Quan X, Song L, Zheng X, Liu S, Ding H, Li S, Xu G, Li X, Liu L (2021) Reduction of autophagosome overload attenuates neuronal cell death after traumatic brain injury. Neuroscience 460:107–119. https://doi.org/10.1016/j.neuroscience.2021.02.007
Qureshi HY, Li T, MacDonald R, Cho CM, Leclerc N, Paudel HK (2013) Interaction of 14–3-3zeta with microtubule-associated protein tau within Alzheimer’s disease neurofibrillary tangles. Biochemistry 52:6445–6455. https://doi.org/10.1021/bi400442d
Rahmati S, Abovsky M, Pastrello C, Kotlyar M, Lu R, Cumbaa CA, Rahman P, Chandran V, Jurisica I (2020) pathDIP 4: an extended pathway annotations and enrichment analysis resource for human, model organisms and domesticated species. Nucleic Acids Res 48:D479–D488. https://doi.org/10.1093/nar/gkz989
Raj R, Kaprio J, Korja M, Mikkonen ED, Jousilahti P, Siironen J (2017) Risk of hospitalization with neurodegenerative disease after moderate-to-severe traumatic brain injury in the working-age population: a retrospective cohort study using the Finnish national health registries. PLoS Med 14:e1002316. https://doi.org/10.1371/journal.pmed.1002316
Redell JB, Liu Y, Dash PK (2009) Traumatic brain injury alters expression of hippocampal microRNAs: potential regulators of multiple pathophysiological processes. J Neurosci Res 87:1435–1448. https://doi.org/10.1002/jnr.21945
Redell JB, Moore AN, Ward NH 3rd, Hergenroeder GW, Dash PK (2010) Human traumatic brain injury alters plasma microRNA levels. J Neurotrauma 27:2147–2156. https://doi.org/10.1089/neu.2010.1481
de Rivero Vaccari JP, Lotocki G, Alonso OF, Bramlett HM, Dietrich WD, Keane RW (2009) Therapeutic neutralization of the NLRP1 inflammasome reduces the innate immune response and improves histopathology after traumatic brain injury. J Cereb Blood Flow Metab 29:1251–1261. https://doi.org/10.1038/jcbfm.2009.46
Roberts GW, Gentleman SM, Lynch A, Murray L, Landon M, Graham DI (1994) Beta amyloid protein deposition in the brain after severe head injury: implications for the pathogenesis of Alzheimer’s disease. J Neurol Neurosurg Psychiatry 57:419–425. https://doi.org/10.1136/jnnp.57.4.419
Rouillard AD, Gundersen GW, Fernandez NF, Wang Z, Monteiro CD, McDermott MG, Ma’ayan A (2016) The harmonizome: a collection of processed datasets gathered to serve and mine knowledge about genes and proteins. Database (oxford). https://doi.org/10.1093/database/baw100
Rui Q, Ni H, Gao F, Dang B, Li D, Gao R, Chen G (2018) LRRK2 contributes to secondary brain injury through a p38/drosha Signaling Pathway After Traumatic Brain Injury in Rats. Front Cell Neurosci 12:51. https://doi.org/10.3389/fncel.2018.00051
Russo MV, McGavern DB (2016) Inflammatory neuroprotection following traumatic brain injury. Science 353:783–785. https://doi.org/10.1126/science.aaf6260
Salehi A, Jullienne A, Baghchechi M, Hamer M, Walsworth M, Donovan V, Tang J, Zhang JH, Pearce WJ, Obenaus A (2018) Up-regulation of Wnt/beta-catenin expression is accompanied with vascular repair after traumatic brain injury. J Cereb Blood Flow Metab 38:274–289. https://doi.org/10.1177/0271678X17744124
Santacruz CA, Vincent JL, Bader A, Rincon-Gutierrez LA, Dominguez-Curell C, Communi D, Taccone FS (2021) Association of cerebrospinal fluid protein biomarkers with outcomes in patients with traumatic and non-traumatic acute brain injury: systematic review of the literature. Crit Care 25:278. https://doi.org/10.1186/s13054-021-03698-z
Satoh J, Kino Y, Niida S (2015) MicroRNA-Seq data analysis pipeline to identify blood biomarkers for Alzheimer’s disease from Public Data. Biomark Insights 10:21–31. https://doi.org/10.4137/BMI.S25132
Schindler CR, Woschek M, Vollrath JT, Kontradowitz K, Lustenberger T, Stormann P, Marzi I, Henrich D (2020) miR-142-3p expression is predictive for severe traumatic brain injury (TBI) in trauma patients. Int J Mol Sci 21:5381. https://doi.org/10.3390/ijms21155381
Schulz J, Takousis P, Wohlers I, Itua IOG, Dobricic V, Rucker G, Binder H, Middleton L, Ioannidis JPA, Perneczky R, Bertram L, Lill CM (2019) Meta-analyses identify differentially expressed micrornas in Parkinson’s disease. Ann Neurol 85:835–851. https://doi.org/10.1002/ana.25490
Sebastiani A, Golz C, Sebastiani PG, Bobkiewicz W, Behl C, Mittmann T, Thal SC, Engelhard K (2017) Sequestosome 1 deficiency delays, but does not prevent brain damage formation following acute brain injury in adult mice. Front Neurosci 11:678. https://doi.org/10.3389/fnins.2017.00678
Sharma A, Chandran R, Barry ES, Bhomia M, Hutchison MA, Balakathiresan NS, Grunberg NE, Maheshwari RK (2014) Identification of serum microRNA signatures for diagnosis of mild traumatic brain injury in a closed head injury model. PLoS ONE 9:e112019. https://doi.org/10.1371/journal.pone.0112019
Sheinerman KS, Toledo JB, Tsivinsky VG, Irwin D, Grossman M, Weintraub D, Hurtig HI, Chen-Plotkin A, Wolk DA, McCluskey LF, Elman LB, Trojanowski JQ, Umansky SR (2017) Circulating brain-enriched microRNAs as novel biomarkers for detection and differentiation of neurodegenerative diseases. Alzheimers Res Ther 9:89. https://doi.org/10.1186/s13195-017-0316-0
Shirdel EA, Xie W, Mak TW, Jurisica I (2011) NAViGaTing the micronome–using multiple microRNA prediction databases to identify signalling pathway-associated microRNAs. PLoS ONE 6:e17429. https://doi.org/10.1371/journal.pone.0017429
Sil S, Periyasamy P, Thangaraj A, Chivero ET, Buch S (2018) PDGF/PDGFR axis in the neural systems. Mol Aspects Med 62:63–74. https://doi.org/10.1016/j.mam.2018.01.006
Smith DH, Johnson VE, Stewart W (2013) Chronic neuropathologies of single and repetitive TBI: substrates of dementia? Nat Rev Neurol 9:211–221. https://doi.org/10.1038/nrneurol.2013.29
Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y, Kaplan S, Dahary D, Warshawsky D, Guan-Golan Y, Kohn A, Rappaport N, Safran M, Lancet D (2016) The genecards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 54:1. https://doi.org/10.1002/cpbi.5
Su EJ, Fredriksson L, Kanzawa M, Moore S, Folestad E, Stevenson TK, Nilsson I, Sashindranath M, Schielke GP, Warnock M, Ragsdale M, Mann K, Lawrence AL, Medcalf RL, Eriksson U, Murphy GG, Lawrence DA (2015) Imatinib treatment reduces brain injury in a murine model of traumatic brain injury. Front Cell Neurosci 9:385. https://doi.org/10.3389/fncel.2015.00385
Sun D, Bullock MR, Altememi N, Zhou Z, Hagood S, Rolfe A, McGinn MJ, Hamm R, Colello RJ (2010) The effect of epidermal growth factor in the injured brain after trauma in rats. J Neurotrauma 27:923–938. https://doi.org/10.1089/neu.2009.1209
Sun L, Liu A, Zhang J, Ji W, Li Y, Yang X, Wu Z, Guo J (2018) miR-23b improves cognitive impairments in traumatic brain injury by targeting ATG12-mediated neuronal autophagy. Behav Brain Res 340:126–136. https://doi.org/10.1016/j.bbr.2016.09.020
Sun C, Liu J, Duan F, Cong L, Qi X (2022) The role of the microRNA regulatory network in Alzheimer’s disease: a bioinformatics analysis. Arch Med Sci 18:206–222. https://doi.org/10.5114/aoms/80619
Tan L, Yu JT, Tan MS, Liu QY, Wang HF, Zhang W, Jiang T, Tan L (2014) Genome-wide serum microRNA expression profiling identifies serum biomarkers for Alzheimer’s disease. J Alzheimers Dis 40:1017–1027. https://doi.org/10.3233/JAD-132144
Tang YL, Fang LJ, Zhong LY, Jiang J, Dong XY, Feng Z (2020) Hub genes and key pathways of traumatic brain injury: bioinformatics analysis and in vivo validation. Neural Regen Res 15:2262–2269. https://doi.org/10.4103/1673-5374.284996
Teasdale G, Jennett B (1974) Assessment of coma and impaired consciousness. Practical Scale Lancet 2:81–84. https://doi.org/10.1016/s0140-6736(74)91639-0
Teasdale G, Jennett B (1976) Assessment and prognosis of coma after head injury. Acta Neurochir (wien) 34:45–55. https://doi.org/10.1007/BF01405862
Thangavelu B, Wilfred BS, Johnson D, Gilsdorf JS, Shear DA, Boutte AM (2020) Penetrating ballistic-like brain injury leads to microRNA dysregulation, BACE1 upregulation, and amyloid precursor protein loss in lesioned rat brain tissues. Front Neurosci 14:915. https://doi.org/10.3389/fnins.2020.00915
Toffolo K, Osei J, Kelly W, Poulsen A, Donahue K, Wang J, Hunter M, Bard J, Wang J, Poulsen D (2019) Circulating microRNAs as biomarkers in traumatic brain injury. Neuropharmacology 145:199–208. https://doi.org/10.1016/j.neuropharm.2018.08.028
Tokar T, Pastrello C, Rossos AEM, Abovsky M, Hauschild AC, Tsay M, Lu R, Jurisica I (2018) mirDIP 4.1-integrative database of human microRNA target predictions. Nucleic Acids Res 46:D360–D370. https://doi.org/10.1093/nar/gkx1144
Tominaga K, Suzuki HI (2019) TGF-beta signaling in cellular senescence and aging-related pathology. Int J Mol Sci. https://doi.org/10.3390/ijms20205002
Villapol S, Wang Y, Adams M, Symes AJ (2013) Smad3 deficiency increases cortical and hippocampal neuronal loss following traumatic brain injury. Exp Neurol 250:353–365. https://doi.org/10.1016/j.expneurol.2013.10.008
von Bernhardi R, Eugenin-von Bernhardi L, Eugenin J (2015) Microglial cell dysregulation in brain aging and neurodegeneration. Front Aging Neurosci 7:124. https://doi.org/10.3389/fnagi.2015.00124
Wang JW, Wang HD, Cong ZX, Zhou XM, Xu JG, Jia Y, Ding Y (2014) Puerarin ameliorates oxidative stress in a rodent model of traumatic brain injury. J Surg Res 186:328–337. https://doi.org/10.1016/j.jss.2013.08.027
Wang XY, Ba YC, Xiong LL, Li XL, Zou Y, Zhu YC, Zhou XF, Wang TH, Wang F, Tian HL, Li JT (2015) Endogenous TGFbeta1 plays a crucial role in functional recovery after traumatic brain injury associated with smad3 signal in rats. Neurochem Res 40:1671–1680. https://doi.org/10.1007/s11064-015-1634-x
Wei C, Luo Y, Peng L, Huang Z, Pan Y (2021) Expression of Notch and Wnt/beta-catenin signaling pathway in acute phase severe brain injury rats and the effect of exogenous thyroxine on those pathways. Eur J Trauma Emerg Surg 47:2001–2015. https://doi.org/10.1007/s00068-020-01359-4
Wei C, Luo Y, Peng L, Huang Z, Pan Y (2020) Expression of Notch and Wnt/beta-catenin signaling pathway in acute phase severe brain injury rats and the effect of exogenous thyroxine on those pathways. Eur J Trauma Emerg Surg. https://doi.org/10.1007/s00068-020-01359-4
Weisz HA, Kennedy D, Widen S, Spratt H, Sell SL, Bailey C, Sheffield-Moore M, DeWitt DS, Prough DS, Levin H, Robertson C, Hellmich HL (2020) MicroRNA sequencing of rat hippocampus and human biofluids identifies acute, chronic, focal and diffuse traumatic brain injuries. Sci Rep 10:3341. https://doi.org/10.1038/s41598-020-60133-z
Wu P, Zhao Y, Haidacher SJ, Wang E, Parsley MO, Gao J, Sadygov RG, Starkey JM, Luxon BA, Spratt H, Dewitt DS, Prough DS, Denner L (2013) Detection of structural and metabolic changes in traumatically injured hippocampus by quantitative differential proteomics. J Neurotrauma 30:775–788. https://doi.org/10.1089/neu.2012.2391
Wurzelmann M, Romeika J, Sun D (2017) Therapeutic potential of brain-derived neurotrophic factor (BDNF) and a small molecular mimics of BDNF for traumatic brain injury. Neural Regen Res 12:7–12. https://doi.org/10.4103/1673-5374.198964
Xie Z, Bailey A, Kuleshov MV, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, Jeon M, Ma’ayan A (2021) Gene set knowledge discovery with enrichr. Curr Protoc 1:e90. https://doi.org/10.1002/cpz1.90
Yan J, Bu X, Li Z, Wu J, Wang C, Li D, Song J, Wang J (2019) Screening the expression of several miRNAs from TaqMan low density array in traumatic brain injury: miR-219a-5p regulates neuronal apoptosis by modulating CCNA2 and CACUL1. J Neurochem 150:202–217. https://doi.org/10.1111/jnc.14717
Yang T, Song J, Bu X, Wang C, Wu J, Cai J, Wan S, Fan C, Zhang C, Wang J (2016) Elevated serum miR-93, miR-191, and miR-499 are noninvasive biomarkers for the presence and progression of traumatic brain injury. J Neurochem 137:122–129. https://doi.org/10.1111/jnc.13534
You WD, Tang QL, Wang L, Lei J, Feng JF, Mao Q, Gao GY, Jiang JY (2016) Alteration of microRNA expression in cerebrospinal fluid of unconscious patients after traumatic brain injury and a bioinformatic analysis of related single nucleotide polymorphisms. Chin J Traumatol 19:11–15. https://doi.org/10.1016/j.cjtee.2016.01.004
Zetterberg H, Winblad B, Bernick C, Yaffe K, Majdan M, Johansson G, Newcombe V, Nyberg L, Sharp D, Tenovuo O, Blennow K (2019) Head trauma in sports—clinical characteristics, epidemiology and biomarkers. J Intern Med 285:624–634. https://doi.org/10.1111/joim.12863
Zhang Z, Yan R, Zhang Q, Li J, Kang X, Wang H, Huan L, Zhang L, Li F, Yang S, Zhang J, Ren X, Yang X (2014) Hes1, a Notch signaling downstream target, regulates adult hippocampal neurogenesis following traumatic brain injury. Brain Res 1583:65–78. https://doi.org/10.1016/j.brainres.2014.07.037
Zhou Q, Yin J, Wang Y, Zhuang X, He Z, Chen Z, Yang X (2021) MicroRNAs as potential biomarkers for the diagnosis of traumatic brain injury: a systematic review and meta-analysis. Int J Med Sci 18:128–136. https://doi.org/10.7150/ijms.48214
Funding
Bioinformatic analyses were in part supported by the Ministry of Education of the Slovak Republic, awarded to BrainTest, s.r.o. within the research scheme “Stimuli for research and development” and research grants VEGA 2/0118/19, VEGA 2/0153/22, APVV-17-0668, APVV-19-0568, APVV-20-0615, and ERA-NET Neuron Neu-Vasc, CIHR-Eranet Neuron (NOD-168163), Ontario Research Fund (RDI 34876), and Canada Foundation for Innovation (CFI 29272, 225404, 33536).
Author information
Authors and Affiliations
Contributions
KM, NC, and SP collected and curated the data. YN and IJ performed the bioinformatic analysis. MC and KM wrote the manuscript. PF, AT, MM, and IJ reviewed and edited the manuscript. All authors read and approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
Authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
10571_2022_1254_MOESM1_ESM.png
Supplementary file1 (PNG 20240 kb)—Target genes for deregulated miRNAs in serum and CSF after sTBI, as identified in mirDIP. Node color for genes represents Gene ontology (GO) molecular function, and node outline for miRNAs highlights experimental evidence, as per legend. Listed gene names are targeted by most of the deregulated miRNAs, and names with green background highlight genes regulated by validated miRNAs
10571_2022_1254_MOESM2_ESM.png
Supplementary file2 (PNG 5614 kb)—Target genes for deregulated miRNAs in plasma and CSF after sTBI, as identified in mirDIP. Node color for genes represents Gene ontology (GO) molecular function, and node outline for miRNAs highlights experimental evidence, as per legend. Listed gene names are targeted by most of the deregulated miRNAs, and names with green background highlight genes regulated by validated miRNAs
10571_2022_1254_MOESM3_ESM.png
Supplementary file3 (PNG 12328 kb)—Target genes for deregulated miRNAs in serum and plasma after sTBI, as identified in mirDIP. Node color for genes represents Gene ontology (GO) molecular function, and node outline for miRNAs highlights experimental evidence, as per legend. Listed gene names are targeted by most of the deregulated miRNAs, and names with green background highlight genes regulated by validated miRNAs
Rights and permissions
About this article
Cite this article
Cente, M., Matyasova, K., Csicsatkova, N. et al. Traumatic MicroRNAs: Deconvolving the Signal After Severe Traumatic Brain Injury. Cell Mol Neurobiol 43, 1061–1075 (2023). https://doi.org/10.1007/s10571-022-01254-z
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10571-022-01254-z