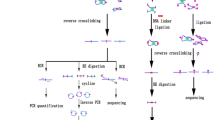
Abstract
Hi-C is a commonly used technology in 3D genomics which can depict global chromatin interactions across eukaryotic genome. Integrating with different datasets, it can also be applied to studying various biological questions, such as nuclear organization, gene transcription regulation, spatiotemporal development, genome assembly, and cancer genomics. During the last decade, the development and application of Hi-C have dramatically changed the view of genome architecture, chromatin conformation, and gene interaction. So far, Hi-C-related studies remain vivacious and controversial; thus, a unified standard of library construction and bioinformatics analysis are urgently needed. In this review, we have summarized its history, development, methodologies, advances, applications, shortages, and future perspectives. We discuss a few limitations of the current Hi-C technologies and future directions for improvement and highlight how Hi-C can bridge 3D structure to gene function. This review will be helpful for scientists who want to engage in the 3D genomics field; it also shows some future tracks.






Similar content being viewed by others
References
Ay F, Bailey TL, Noble WS. Statistical confidence estimation for Hi-C data reveals regulatory chromatin contacts. Genome Res. 2014;24(6):999–1011.
Battulin NR, Fishman VS, Mazur AM, Pomaznoy M, Khabarova AA, Afonnikov DA, et al. Comparison of the three-dimensional organization of sperm and fibroblast genomes using the Hi-C approach. Genome Biol. 2015;16(1):77.
Belton J, Mccord RP, Gibcus JH, Naumova N, Zhan Y, Dekker J. Hi–C: a comprehensive technique to capture the conformation of genomes. Methods. 2012;58(3):268–76.
Berlivet S, Paquette D, Dumouchel A, Langlais D, Dostie J, Kmita M. Clustering of tissue-specific sub-TADs accompanies the regulation of HoxA genes in developing limbs. PLoS Genet. 2013;9(12):e1004018.
Bickhart DM, Rosen BD, Koren S, Sayre BL, Hastie AR, Chan S, et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat Genet. 2017;49(4):643–50.
Bintu B, Mateo LJ, Su J-H, Sinnott-Armstrong NA, Parker M, Kinrot S, et al. Super-resolution chromatin tracing reveals domains and cooperative interactions in single cells. Science. 2018;362(6413):eaau1783. https://doi.org/10.1126/science.aau1783.
Bonev B, Cavalli G. Organization and function of the 3D genome. Nat Rev Genet. 2016;17(11):661–78.
Burton JN, Adey A, Patwardhan RP, Qiu R, Kitzman JO, Shendure J. Chromosome-scale scaffolding of de novo genome assemblies based on chromatin interactions. Nat Biotechnol. 2013;31(12):1119–25.
Cavalli G. Chromosomes: now in 3D! Nat Rev Mol Cell Biol. 2014;15(1):6–6.
Chambers EV, Bickmore WA, Semple CAM. Divergence of mammalian higher order chromatin structure is associated with developmental loci. PLoS Comput Biol. 2013;9(4):e1003017.
Chen H, Li C, Peng X, Zhou Z, Weinstein JN, Liang H. A pan-cancer analysis of enhancer expression in nearly 9000 patient samples. Cell. 2018a;173(2):386–399.e12.
Chen F, Li G, Zhang MQ, Chen Y. HiCDB: a sensitive and robust method for detecting contact domain boundaries. Nucleic Acids Research, gky789-gky789. 2018b. https://doi.org/10.1093/nar/gky789.
Cremer T, Cremer M, Cremer C. The 4D nucleome: genome compartmentalization in an evolutionary context. Biochemistry. 2018;83(4):313–25.
De Laat W, Dekker J. 3C-based technologies to study the shape of the genome. Methods. 2012;58(3):189–91.
Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295(5558):1306–11.
Dekker J, Belmont AS, Guttman M, Leshyk VO, Lis JT, Lomvardas S, et al. The 4D nucleome project. Nature. 2017;549(7671):219–26.
Diament A, Tuller T. Tracking the evolution of 3D gene organization demonstrates its connection to phenotypic divergence. Nucleic Acids Res. 2017;45(8):4330–43.
Diament A, Tuller T. Modeling three-dimensional genomic organization in evolution and pathogenesis. Semin Cell Dev Biol. 2018. https://doi.org/10.1016/j.semcdb.2018.07.008.
Diament A, Pinter RY, Tuller T. Three-dimensional eukaryotic genomic organization is strongly correlated with codon usage expression and function. Nat Commun. 2014;5(1):5876.
Dixon JR, Selvaraj S, Yue F, Kim A, Li Y, Shen Y, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature. 2012;485(7398):376–80.
Dixon JR, Jung I, Selvaraj S, Shen Y, Antosiewiczbourget J, Lee AY, et al. Chromatin architecture reorganization during stem cell differentiation. Nature. 2015;518(7539):331–6.
Dixon JR, Gorkin DU, Ren B. Chromatin domains: the unit of chromosome organization. Mol Cell. 2016;62(5):668–80.
Dogan ES, Liu C. Three-dimensional chromatin packing and positioning of plant genomes[J]. Nature plants. 2018; 4(8),521–529, doi:https://doi.org/10.1038/s41477-018-0199-5.
Dostie J, Richmond T, Arnaout R, Selzer RR, Lee W, Honan T, et al. Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16(10):1299–309.
Du Z, Zheng H, Huang B, Ma R, Wu J, Zhang X, et al. Allelic reprogramming of 3D chromatin architecture during early mammalian development. Nature. 2017;547(7662):232–5.
Du X, Huang G, He S, Yang Z, Sun G, Ma X, et al. Resequencing of 243 diploid cotton accessions based on an updated A genome identifies the genetic basis of key agronomic traits. Nat Genet. 2018;50(6):796–802. https://doi.org/10.1038/s41588-018-0116-x.
Duan Z, Blau CA. The genome in space and time. BioEssays. 2012;34(9):800–10.
Dudchenko O, Batra SS, Omer AD, Nyquist SK, Hoeger M, Durand NC, et al. De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science. 2017;356(6333):92–5.
Durand NC, Robinson JT, Shamim MS, Machol I, Mesirov JP, Lander ES, et al. Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst. 2016;3(1):99–101.
Fang R, Yu M, Li G, Chee S, Liu T, Schmitt AD, et al. Mapping of long-range chromatin interactions by proximity ligation-assisted ChIP-seq. Cell Res. 2016;26(12):1345–8.
Feingold EA, Good PJ, Guyer MS, Kamholz S, Liefer L, Wetterstrand KA, et al. The ENCODE (ENCyclopedia of DNA Elements) project. Science. 2004;306(5696):636–40.
Flavahan WA, Drier Y, Liau BB, Gillespie SM, Venteicher AS, Stemmerrachamimov A, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature. 2016;529(7584):110–4.
Flyamer IM, Gassler J, Imakaev M, Brandao HB, Ulianov SV, Abdennur N, et al. Single-nucleus Hi-C reveals unique chromatin reorganization at oocyte-to-zygote transition. Nature. 2017;544(7648):110–4.
Fullwood MJ, Liu MH, Pan YF, Liu J, Xu H, Mohamed YB, et al. An oestrogen-receptor-α-bound human chromatin interactome. Nature. 2009;462(7269):58–64.
Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell. 2013;49(5):773–82.
Giorgetti L, Lajoie BR, Carter AC, Attia M, Zhan Y, Xu J, et al. Structural organization of the inactive X chromosome in the mouse. Nature. 2016;535(7613):575–9.
Goetze S, Mateoslangerak J, Van Driel R. Three-dimensional genome organization in interphase and its relation to genome function. Semin Cell Dev Biol. 2007;18(5):707–14.
Grosberg AY, Nechaev SK, Shakhnovich EI. The role of topological constraints in the kinetics of collapse of macromolecules. J Phys. 1988;49(12):2095–100.
Grosberg A, Rabin Y, Havlin S, Neer A. Crumpled globule model of the three-dimensional structure of DNA. Epl. 1993;23(5):373–8.
Heinz S, Benner C, Spann NJ, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–89.
Heinz S, Texari L, Hayes MGB, Urbanowski M, Chang MW, Givarkes N, et al. Transcription elongation can affect genome 3D structure. Cell. 2018;174(6):1522–1536.e1522. https://doi.org/10.1016/j.cell.2018.07.047.
Ho JWK, Jung YL, Liu T, Alver BH, Lee S, Ikegami K, et al. Comparative analysis of metazoan chromatin organization. Nature. 2014;512(7515):449–52.
Hu M, Deng K, Qin Z, Liu JS. Understanding spatial organizations of chromosomes via statistical analysis of Hi-C data. Quant Biol. 2013;1(2):156–74.
Hug CB, Grimaldi AG, Kruse K, Vaquerizas JM. Chromatin architecture emerges during zygotic genome activation independent of transcription. Cell. 2017;169(2):216–28.
Ibnsalem J, Muro EM, Andradenavarro MA. Co-regulation of paralog genes in the three-dimensional chromatin architecture. Nucleic Acids Res. 2017;45(1):81–91.
Javierre B, Burren OS, Wilder SP, Kreuzhuber R, Hill SM, Sewitz S, et al. Lineage-specific genome architecture links enhancers and non-coding disease variants to target gene promoters. Cell. 2016;167(5):1369–84.
Jin F, Li Y, Dixon JR, Selvaraj S, Ye Z, Lee AY, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature. 2013;503(7475):290–4.
Kalhor R, Tjong H, Jayathilaka N, Alber F, Chen L. Genome architectures revealed by tethered chromosome conformation capture and population-based modeling. Nat Biotechnol. 2012;30(1):90–8.
Ke Y, Xu Y, Chen XW, Feng S, Liu Z, Sun Y, et al. 3D chromatin structures of mature gametes and structural reprogramming during mammalian embryogenesis. Cell. 2017;170(2):367–81.
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T. Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev Genet. 2007;8(2):104–15.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–9.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754–60.
Li R, Liu Y, Li T, Li C. 3Disease browser: a web server for integrating 3D genome and disease-associated chromosome rearrangement data. Sci Rep. 2016;6(1):34651.
Li G, Chen Y, Snyder MP, Zhang MQ. ChIA-PET2: a versatile and flexible pipeline for ChIA-PET data analysis. Nucleic Acids Res. 2017;45(1):e4.
Li T, Jia L, Cao Y, Chen Q, Li C. OCEAN-C: mapping hubs of open chromatin interactions across the genome reveals gene regulatory networks[J]. Genome Biol. 2018a;19(1). https://doi.org/10.1186/s13059-018-1430-4.
Li R, Liu Y, Hou Y, Gan J, Wu P, Li C. 3D genome and its disorganization in diseases. Cell Biol Toxicol. 2018b; 1–15.
Liang Z, Li G, Wang Z, Djekidel MN, Li Y, Qian M, et al. BL-Hi-C is an efficient and sensitive approach for capturing structural and regulatory chromatin interactions. Nat Commun. 2017;8(1):1622.
Lieberman-Aiden E, Van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, et al. Comprehensive mapping of long range interactions reveals folding principles of the human genome. Science. 2009;326(5950):289–93.
Lin D, Hong P, Zhang S, Xu W, Jamal M, Yan K, et al. Digestion-ligation-only Hi-C is an efficient and cost-effective method for chromosome conformation capture[J]. Nat Genet. 2018;50(5), 754–763. https://doi.org/10.1038/s41588-018-0111-2.
Liu X, Zhang Y, Chen Y, Li M, Zhou F, Li K, et al. In situ capture of chromatin interactions by biotinylated dCas9. Cell. 2017;170(5):1028–43.
Long HK, Prescott SL, Wysocka J. Ever-changing landscapes: transcriptional enhancers in development and evolution. Cell. 2016;167(5):1170–87.
Ma W, Ay F, Lee C, Gulsoy G, Deng X, Cook S, et al. Fine-scale chromatin interaction maps reveal the cis-regulatory landscape of human lincRNA genes. Nat Methods. 2015;12(1):71–8.
Ma T, Chen L, Shi M, Niu J, Zhang X, Yang X, et al. Developing novel methods to image and visualize 3D genomes[J]. Cell Biol Toxicol. 2018;34(5):367–380. https://doi.org/10.1007/s10565-018-9427-z.
Mascher M, Gundlach H, Himmelbach A, Beier S, Twardziok SO, Wicker T, et al. A chromosome conformation capture ordered sequence of the barley genome. Nature. 2017;544(7651):427–33.
Mateoslangerak J, Bohn M, De Leeuw WC, Giromus O, Manders EMM, Verschure PJ, et al. Spatially confined folding of chromatin in the interphase nucleus. Proc Natl Acad Sci U S A. 2009;106(10):3812–7.
Merelli I, Tordini F, Drocco M, Aldinucci M, Lio P, Milanesi L. Integrating multi-omic features exploiting chromosome conformation capture data. Front Genet. 2015;6(40):40.
Mifsud B, Tavarescadete F, Young AN, Sugar R, Schoenfelder S, Ferreira L, et al. Mapping long-range promoter contacts in human cells with high-resolution capture Hi-C. Nat Genet. 2015;47(6):598–606.
Mishra A, Hawkins RD. Three-dimensional genome architecture and emerging technologies: looping in disease. Genome Med. 2017;9(1):87.
Mumbach MR, Rubin AJ, Flynn RA, Dai C, Khavari PA, Greenleaf WJ, et al. HiChIP: efficient and sensitive analysis of protein-directed genome architecture[J]. Nat Methods. 2016;13(11):919–922. https://doi.org/10.1038/nmeth.3999.
Munkel C, Langowski J. Chromosome structure predicted by A polymer model. Phys Rev E. 1998;57(5):5888–96.
Nagano T, Lubling Y, Stevens TJ, Schoenfelder S, Yaffe E, Dean W, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature. 2013;502(7469):59–64.
Nagano T, Lubling Y, Yaffe E, Wingett SW, Dean W, Tanay A, et al. Single-cell Hi-C for genome-wide detection of chromatin interactions that occur simultaneously in a single cell. Nat Protoc. 2015a;10(12):1986–2003.
Nagano T, Varnai C, Schoenfelder S, Javierre B, Wingett SW, Fraser P. Comparison of Hi-C results using in-solution versus in-nucleus ligation. Genome Biol. 2015b;16(1):175.
Nagano T, Lubling Y, Varnai C, Dudley C, Leung W, Baran Y, et al. Cell-cycle dynamics of chromosomal organization at single-cell resolution. Nature. 2017;547(7661):61–7.
Novo CL, Javierre B, Cairns J, Segondspichon A, Wingett SW, Freirepritchett P, et al. Long-range enhancer interactions are prevalent in mouse embryonic stem cells and are reorganized upon pluripotent state transition. Cell Rep. 2018;22(10):2615–27.
Ou HD, Phan S, Deerinck TJ, Thor A, Ellisman MH, Shea CCO. ChromEMT: visualizing 3D chromatin structure and compaction in interphase and mitotic cells. Science. 2017;357(6349):eaag0025.
Pancaldi V, Carrillodesantapau E, Javierre BM, Juan D, Fraser P, Spivakov M, et al. Integrating epigenomic data and 3D genomic structure with a new measure of chromatin assortativity. Genome Biol. 2016;17(1):152.
Papantonis A, Cook PR. Genome architecture and the role of transcription. Curr Opin Cell Biol. 2010;22(3):271–6.
Perichupkes D, Meuleman W, Pagie L, Bruggeman SWM, Solovei I, Brugman W, et al. Molecular maps of the reorganization of genome-nuclear lamina interactions during differentiation. Mol Cell. 2010;38(4):603–13.
Phillipscremins JE, Sauria ME, Sanyal A, Gerasimova T, Lajoie BR, Bell JSK, et al. Architectural protein subclasses shape 3D organization of genomes during lineage commitment. Cell. 2013;153(6):1281–95.
Quinodoz SA, Ollikainen N, Tabak B, Palla A, Schmidt JM, Detmar E, et al. Higher-order inter-chromosomal hubs shape 3D genome organization in the nucleus. Cell. 2018;174:744–757.e24. https://doi.org/10.1016/j.cell.2018.05.024.
Ramani V, Cusanovich DA, Hause RJ, Ma W, Qiu R, Deng X, et al. Mapping 3D genome architecture through in situ DNase Hi-C. Nat Protoc. 2016;11(11):2104–21.
Ramani V, Deng X, Qiu R, Gunderson KL, Steemers FJ, Disteche CM, et al. Massively multiplex single-cell Hi-C. Nat Methods. 2017;14(3):263–6.
Rao SSP, Huntley M, Durand NC, Stamenova EK, Bochkov ID, Robinson JT, et al. A 3D map of the human genome at kilobase resolution reveals principles of chromatin looping. Cell. 2014;159(7):1665–80.
Remeseiro S, Hornblad A, Spitz F. Gene regulation during development in the light of topologically associating domains. Wiley Interdiscip Rev Dev Biol. 2016;5(2):169–85.
Rowley MJ, Nichols MH, Lyu X, Andokuri M, Ism R, Hermetz K, et al. Evolutionarily conserved principles predict 3D chromatin organization. Mol Cell. 2017;67(5):837–852.e7.
Rudan MV, Barrington C, Henderson S, Ernst C, Odom DT, Tanay A, et al. Comparative Hi-C reveals that CTCF underlies evolution of chromosomal domain architecture. Cell Rep. 2015;10(8):1297–309.
Schmitt AD, Hu M, Jung I, Xu Z, Qiu Y, Tan CL, et al. A compendium of chromatin contact maps reveals spatially active regions in the human genome. Cell Rep. 2016a;17(8):2042–59.
Schmitt AD, Hu M, Ren B. Genome-wide mapping and analysis of chromosome architecture. Nat Rev Mol Cell Biol. 2016b;17(12):743–55.
Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat Genet. 2010;42(1):53–61.
Selvaraj S, Dixon JR, Bansal V, Ren B. Whole-genome haplotype reconstruction using proximity-ligation and shotgun sequencing. Nat Biotechnol. 2013;31(12):1111–8.
Servant N, Varoquaux N, Lajoie BR, Viara E, Chen C, Vert J, et al. HiC-Pro: an optimized and flexible pipeline for Hi-C data processing. Genome Biol. 2015;16(1):259.
Sexton T, Cavalli G. The role of chromosome domains in shaping the functional genome. Cell. 2015;160(6):1049–59.
Sexton T, Yaffe E, Kenigsberg E, Bantignies F, Leblanc B, Hoichman M, et al. Three-dimensional folding and functional organization principles of the Drosophila genome. Cell. 2012;148(3):458–72.
Shao Y, Lu N, Wu Z, Cai C, Wang S, Zhang LL, et al. Creating a functional single-chromosome yeast. Nature. 2018;560:331–5.
Simonis M, Klous P, Splinter E, Moshkin YM, Willemsen R, De Wit E, et al. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat Genet. 2006;38(11):1348–54.
Smallwood A, Ren B. Genome organization and long-range regulation of gene expression by enhancers. Curr Opin Cell Biol. 2013;25(3):387–94.
Stadhouders R, Vidal E, Serra F, Stefano BD, Dily FL, Quilez J, et al. Transcription factors orchestrate dynamic interplay between genome topology and gene regulation during cell reprogramming. Nat Genet. 2018;50(2):238–49.
Stamatoyannopoulos JA, Snyder M, Hardison RC, Ren B, Gingeras TR, Gilbert DM, et al. An encyclopedia of mouse DNA elements (mouse ENCODE). Genome Biol. 2012;13(8):418.
Stevens TJ, Lando D, Basu S, Atkinson L, Cao Y, Lee SF, et al. 3D structures of individual mammalian genomes studied by single-cell Hi-C. Nature. 2017;544(7648):59–64.
Szalaj P, Plewczynski D. Three-dimensional organization and dynamics of the genome[J]. Cell Biol Toxicol. 2018;34:381–404. https://doi.org/10.1007/s10565-018-9428-y.
Tan L, Xing D, Chang C, Li H, Xie X. Three-dimensional genome structures of single diploid human cells. Science. 2018;361(6405):924–8.
Ulianov SV, Tachibanakonwalski K, Razin SV. Single-cell Hi-C bridges microscopy and genome-wide sequencing approaches to study 3D chromatin organization. BioEssays. 2017;39(10):1700104.
Uuskulareimand L, Hou H, Samavarchitehrani P, Rudan MV, Liang M, Medinarivera A, et al. Topoisomerase II beta interacts with cohesin and CTCF at topological domain borders. Genome Biol. 2016;17(1):182.
Walter J, Schermelleh L, Cremer M, Tashiro S, Cremer T. Chromosome order in HeLa cells changes during mitosis and early G1, but is stably maintained during subsequent interphase stages. J Cell Biol. 2003;160(5):685–97.
Wang Y, Fan C, Zheng Y, Li C. Dynamic chromatin accessibility modeled by Markov process of randomly-moving molecules in the 3D genome. Nucleic Acids Res. 2017;45(10):85.
Wang Y, Song F, Zhang B, Zhang L, Xu J, Kuang D, et al. The 3D genome browser: a web-based browser for visualizing 3D genome organization and long-range chromatin interactions. [journal article]. Genome Biology. 2018;19(1):151. https://doi.org/10.1186/s13059-018-1519-9.
Wingett SW, Ewels P, Furlanmagaril M, Nagano T, Schoenfelder S, Fraser P, et al. HiCUP: pipeline for mapping and processing Hi-C data. F1000Research. 2015;4:1310.
Yu M, Ren B. The three-dimensional organization of mammalian genomes. Annu Rev Cell Dev Biol. 2017;33(1):265–89.
Yue F, Cheng Y, Breschi A, Vierstra J, Wu W, Ryba T, et al. A comparative encyclopedia of DNA elements in the mouse genome. Nature. 2014;515(7527):355–64.
Zhan Y, Mariani L, Barozzi I, Schulz EG, Blüthgen N, Stadler M, et al. Reciprocal insulation analysis of Hi-C data shows that TADs represent a functionally but not structurally privileged scale in the hierarchical folding of chromosomes. Genome Res. 2017;27(3):479–90.
Zhang Y, Xiang Y, Yin Q, Du Z, Peng X, Wang Q, et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat Genet. 2018;50(1):96–105.
Zhao Z, Tavoosidana G, Sjolinder M, Gondor A, Mariano P, Wang S, et al. Circular chromosome conformation capture (4C) uncovers extensive networks of epigenetically regulated intra- and interchromosomal interactions. Nat Genet. 2006;38(11):1341–7.
Zheng M, Tian SZ, Maurya R, Lee B, Kim M, Capurso D, et al. Multiplex chromatin interaction analysis with single-molecule precision. bioRxiv. 2018. https://doi.org/10.1101/252049.
Zhou X, Lowdon RF, Li D, Lawson HA, Madden PAF, Costello JF, et al. Exploring long-range genome interactions using the WashU epigenome browser. Nat Methods. 2013;10(5):375–6.
Acknowledgements
The present paper was supported by the Thousand Talents Plan for Young Professionals (Y.Z.), the Agricultural Science and Technology Innovation Program, the Fundamental Research Funds for Central Non-profit Scientific Institution (Y2017CG26), the Agricultural Science and Technology Innovation Program Cooperation and Innovation Mission (CAAS-XTCX2016001-3), and the Elite Young Scientists Program of CAAS (CAASQNYC-KYYJ-41).
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Rights and permissions
About this article
Cite this article
Kong, S., Zhang, Y. Deciphering Hi-C: from 3D genome to function. Cell Biol Toxicol 35, 15–32 (2019). https://doi.org/10.1007/s10565-018-09456-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10565-018-09456-2