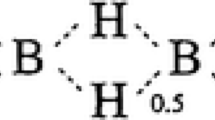
Abstract
The main methods of molecular modeling and specialized software for the development and analysis of molecular models are considered. In particular, the article describes the results of the first stage of developing an environment for the analysis of the molecular and biomolecular interaction that is based on the formalism of behavior algebra and insertion modeling. The results of the experiment of applying the proposed approach to modeling the covalent nonpolar bond are given.
Similar content being viewed by others
References
A. Letichevsky, O. Letychevskyi, and V. Peschanenko, “Insertion modeling and its applications,” Comput. Sci. J. Mold., Vol. 24, No. 3 (72), 357–370 (2016).
T. Lengauer and M. Rarey, “Computational methods for biomolecular docking,” Current Opinion in Structural Biology, Vol. 6, No. 3, 402–406 (1996). https://doi.org/10.1016/s0959-440x(96)80061-3.
DOCK. URL: http://dock.compbio.ucsf.edu.
GOLD. URL: http://www.ccdc.cam.ac.uk/products/life_sciences/gold/.
FLEXX. URL: http://www.biosolveit.de/FlexX/.
FRED. URL: http://www.eyesopen.com/products/applications/fred.html.
AUTODOCK. URL: http://autodock.scripps.edu.
DOCKINGSHOP. URL: http://vis.lbl.gov/~scrivelli/Publicsilvia_page/DockingShop/html.
J. M. Haile, Molecular Dynamics Simulation: Elementary Methods, John Wiley & Sons, New York (1992).
D. C. Rapaport, The Art of Molecular Dynamics Simulation, 2nd ed., Cambridge University Press, Cambridge (2004). https://doi.org/10.1017/CBO9780511816581.
K. Binder (ed.), Monte Carlo Methods in Statistical Physics, Springer, Berlin–Heidelberg (1986).
K. Binder and D. W. Heerman, Monte Carlo Simulation in Statistical Physics: An Introduction, Springer-Verlag, Berlin (1992).
N. S. V. Barbosa, E. R. A. Lima, and F. W. Tavares, Molecular Modeling in Chemical Engineering. Elsevier Reference Collection in Chemistry, Molecular Sciences and Chemical Engineering, Elsevier (2017). https://doi.org/10.1016/B978-0-12-409547-2.13915-0.
A. E. Doroshenko, V. D. Havruchenko, V. I. Egorov, and L. N. Suslovà, “The modeling of the results of quantum-chemical calculations in the Visual Quantum System,” Upr. Sist. Mash., No. 5, 83–87 (2012).
J. R. Faeder, M. L. Blinov, and W. S. Hlavacek, “Rule-based modeling of biochemical systems with BioNetGen,” in: I. Maly (ed.), Systems Biology. Methods in Molecular Biology, Vol. 500, Humana Press (2009), pp. 113–167. https://doi.org/10.1007/978-1-59745-525-1_5.
T. Sneha and J. J. Georrge, “In silico protein engineering: Methods and Tools,” in: Proc. of 10th National Sci. Symp. on Recent Trends in Science and Technology, Christ Publications, Gujarat (2018), pp. 73–80.
E. Chovancova, A. Pavelka, P. Benes, O. Strnad, J. Brezovsky, B. Kozlikova, A. Gora, V. Sustr, M. Klvana, P. Medek, L. Biedermannova, J. Sochor, and J. Damborsky, “CAVER 3.0: A tool for the analysis of transport pathways in dynamic protein structures,” PLoS Comput. Biol. Vol. 8(10), e1002708 (2012). https://doi.org/10.1371/journal.pcbi.1002708.
J. Smadbeck, M. B. Peterson, G. A. Khoury, M. S. Taylor, and C. A. Floudas, “Protein WISDOM: A workbench for in silico de novo design of biomolecules,” J. Vis. Exp., Iss. 77, e50476 (2013). https://doi.org/10.3791/50476.
P. Mitra, D. Shultis, and Y. Zhang, “EvoDesign: De novo protein design based on structural and evolutionary profiles,” Nucleic Acids Research, Vol. 41, Iss. W1, W273–W280 (2013). https://doi.org/10.1093/nar/gkt384.
A. Chatterjee, J. Guedj, and A. S. Perelson, “Mathematical modeling of HCV infection: What can it teach us in the era of direct-acting antiviral agents?” Antivir Ther., Vol. 17, 1171–1182 (2012). https://doi.org/10.3851/IMP2428.
The BioSPI PROJECT. URL: http://www.wisdom.weizmann.ac.il/~biospi/index_main.html.
M. R. Lakin, L. Paulevé, and A. Phillips, “Stochastic simulation of multiple process calculi for biology,” Theor. Comput. Sci., Vol. 431, 181–206 (2012). https://doi.org/10.1016/j.tcs.2011.12.057.
Chemcraft. URL: http://www.chemcraftprog.com/ru/.
Avogadro. URL: https://avogadro.ru.malavida.com/#gref.
ChemDrawPro. URL: https://www.cambridgesoft.com/Ensemble_for_Chemistry/ChemDraw/ChemDrawPro.
Home Page for RasMol and OpenRasMol. URL: http://www.openrasmol.org/.
PyMOL. URL: https://pymol.org/2/.
T. D. Goddard, C. C. Huang, E. C. Meng, E. F. Pettersen, G. S. Couch, J. H. Morris, and T. E. Ferrin, “UCSF ChimeraX: Meeting modern challenges in visualization and analysis,” Protein Sci., Special Issue on Tools for Protein Science, Vol. 27, Iss. 1, 14–25 (2018). https://doi.org/10.1002/pro.3235.
M. W. Schmidt, K. K. Baldridge, J. A. Boatz, S. T. Elbert, M. S. Gordon, J. H. Jensen, S. Koseki, N. Matsunaga, K. A. Nguyen, S. Su, T. L. Windus, M. Dupuis, and J. A. Montgomery Jr, “General atomic and molecular electronic structure system,” J. Comput. Chem., Vol. 14, Iss. 11, 1347–1363 (1993). https://doi.org/10.1002/jcc.540141112.
Program for Mathematical Modeling of Kinetics of Complex Chemical Reactions KINET. URL: http://www.icho39.chem.msu.ru/downloads/kinet.pdf.
ISB. URL: http://labs.systemsbiology.net/bolouri/software/Dizzy/.
BOSS (Biochemical and Organic Simulation System). URL: http://zarbi.chem.yale.edu/software.html.
M. L. Blinov, J. R. Faeder, B. Goldstein, W. S. Hlavacek, “BioNetGen: Software for rule-based modeling of signal transduction based on the interactions of molecular domains,” Bioinformatics, Vol. 20, Iss. 17, 3289–3291 (2004). https://doi.org/10.1093/bioinformatics/bth378.
L. A. Harris, J. S. Hogg, J.-J. Tapia, J. A. P. Sekar, S. Gupta, I. Korsunsky, A. Arora, D. Barua, R. P. Sheehan, and J. R. Faeder, “BioNetGen 2.2: Advances in rule-based modeling,” Bioinformatics, Vol. 32, Iss. 21, 3366–3368 (2016). https://doi.org/10.1093/bioinformatics/btw469.
TinkerTools. URL: https://tinkertools.org/.
Spartan (Chemistry Software). URL: https://www.wavefun.com/.
F. Mohamadi, N. G. J. Richards, W. C. Guida, R. Liskamp, M. Lipton, C. Caufield, and W. C. Still, “Macromodel — an integrated software system for modeling organic and bioorganic molecules using molecular mechanics,” J. Comput. Chem., Vol. 11, Iss. 4, 440–467 (1990).
S. Stoma, M. Fröhlich, S. Gerber, and E. Klipp, “STSE: Spatio-temporal simulation environment dedicated to biology,” BMC Bioinformatics, Vol. 12, Article number 126 (2011).
Abalone II. URL: http://www.biomolecular-modeling.com/Abalone/.
Molecular Forecaster. URL: https://www.molecularforecaster.com/.
R. Salomon-Ferrer, D. A. Case, and R. C. Walker, “An overview of the Amber biomolecular simulation package,” WIREs Comput. Mol. Sci., Vol. 3, Iss. 2, 198–210 (2013).
Integrated Computer-Aided Molecular Design Platform. URL: https://www.chemcomp.com/Products.htm.
B. R. Brooks, C. L. Brooks III, A. D. Mackerell Jr, L. Nilsson, R. J. Petrella, B. Roux, Y. Won, G. Archontis, C. Bartels, S. Boresch, A. Caflisch, L. Caves, Q. Cui, A. R. Dinner, M. Feig, S. Fischer, J. Gao, M. Hodoscek, W. Im, K. Kuczera, T. Lazaridis, J. Ma, V. Ovchinnikov, E. Paci, R. W. Pastor, C. B. Post, J. Z. Pu, M. Schaefer, B. Tidor, R. M. Venable, H. L. Woodcock, X. Wu, W. Yang, D. M. York, and M. Karplus, “CHARMM: The biomolecular simulation program,” J. Comput. Chem., Vol. 30, Iss. 10, 1545–1614 (2009). https://doi.org/10.1002/jcc.21287.
APS & IMS. URL: http//www.apsystem.org.ua.
O. Letychevskyi, V. Peschanenko, M. Poltoratskyi, and Y. Tarasich, “Platform for modeling of algebraic behavior: Experience and conclusions,” CEUR Workshop Proc., Vol. 2732, 42–57 (2020).
A. Letichevsky and D. Gilbert, “A model for interaction of agents and environments,” in: D. Bert, C. Choppy, and P. D. Mosses (eds.), Recent Trends in Algebraic Development Techniques, WADT 1999; Lecture Notes in Computer Science, Vol. 1827, Springer, Berlin–Heidelberg (2000), pp. 311–328. https://doi.org/10.1007/978-3-540-44616-3_18.
S. Baranov, C. Jervis, V. Kotlyarov, A. Letichevsky, T. Weigert, “Leveraging UML to deliver correct telecom applications,” in: L. Lavagno, G. Martin, and B. Selic, (eds.), UML for Real: Design of Embedded Real-Time Systems, Amsterdam, Kluwer Acad. Publ. (2003), pp. 323–342.
A. Ad. Letichevsky, Yu. V. Kapitonova, V. A. Volkov, A. A. Letichevsky, S. N. Baranov, V. P. Kotlyarov, and T. Weigert, “System specification with basic protocols,” Cybern. Syst. Analysis, Vol. 41, No. 4, 479–493 (2005). https://doi.org/10.1007/s10559-005-0083-y.
A. Letichevsky, J. Kapitonova, A. Letichevsky Jr, V. Volkov, S. Baranov, V. Kotlyarov, and T. Weigert, “Basic protocols, message sequence charts, and the verification of requirements specifications,” Computer Networks, Vol. 49, Iss. 5, 661–675 (2005).
R. F. W. Bader, Atoms in Molecules: A Quantum Theory, Oxford, Oxford University Press (1990).
P. S. V. Kumar, V. Raghavendra, and V. Subramanian, “Bader’s theory of atoms in molecules (AIM) and its applications to chemical bonding,” J. Chem. Sci., Vol. 128, No. 1, 1527–1536 (2016).
Author information
Authors and Affiliations
Corresponding author
Additional information
Translated from Kibernetyka ta Systemnyi Analiz, No. 3, May–June, 2022, pp. 150–163.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Letychevskyi, O., Volkov, V., Tarasich, Y. et al. Modern Methods and Software Systems of Molecular Modeling and Application of Behavior Algebra. Cybern Syst Anal 58, 454–464 (2022). https://doi.org/10.1007/s10559-022-00482-x
Received:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10559-022-00482-x