Abstract
Background
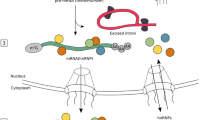
Long non-coding RNAs (lncRNAs) have evolved as a critical regulatory mechanism for almost all biological processes. By dynamically interacting with their molecular partners, lncRNAs regulate gene activity at multiple levels ranging from transcription, pre-mRNA splicing, RNA transporting, RNA decay, and translation of mRNA.
Results and Conclusions
Dysregulation of lncRNAs has been associated with human diseases, including cancer, neurodegenerative, and cardiometabolic diseases. However, as lncRNAs are usually much less conserved than mRNAs at the sequence level, most human lncRNAs are either primate or human specific. The pathophysiological significance of human lncRNAs is still mostly unclear due to the persistent limitations in studying human-specific genes. This review will focus on recent discoveries showing human lncRNAs’ roles in regulating metabolic homeostasis and the potential of targeting this unique group of genes for treatment of cardiometabolic diseases.



Similar content being viewed by others
Data Availability
Not applicable.
Code Availability
Not applicable.
References
Lu W, Cao F, Wang S, Sheng X, Ma J. LncRNAs: the regulator of glucose and lipid metabolism in tumor cells. Front Oncol. 2019;9:1099.
Lin YH. Crosstalk of lncRNA and cellular metabolism and their regulatory mechanism in cancer. Int J Mol Sci. 2020;21(8):2947.
Zhao XY, Lin JD. Long noncoding RNAs: a new regulatory code in metabolic control. Trends Biochem Sci. 2015;40(10):586–96.
Zhang X, Li DY, Reilly MP. Long intergenic noncoding RNAs in cardiovascular diseases: challenges and strategies for physiological studies and translation. Atherosclerosis. 2019;281:180–8.
Muret K, Désert C, Lagoutte L, Boutin M, Gondret F, Zerjal T, et al. Long noncoding RNAs in lipid metabolism: literature review and conservation analysis across species. BMC Genomics. 2019;20(1):882.
Zhao Y, Wu J, Liangpunsakul S, Wang L. Long non-coding RNA in liver metabolism and disease: current status. Liver Res. 2017;1(3):163–7.
Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. 2014;505(7485):635–40.
Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. 2015;11(7):1110–22.
Chen J, Shishkin AA, Zhu X, Kadri S, Maza I, Guttman M, et al. Evolutionary analysis across mammals reveals distinct classes of long non-coding RNAs. Genome Biol. 2016;17:19.
Washietl S, Kellis M, Garber M. Evolutionary dynamics and tissue specificity of human long noncoding RNAs in six mammals. Genome Res. 2014;24(4):616–28.
Halley P, Kadakkuzha BM, Faghihi MA, Magistri M, Zeier Z, Khorkova O, et al. Regulation of the apolipoprotein gene cluster by a long noncoding RNA. Cell Rep. 2014;6(1):222–30.
Ruan X, Li P, Chen Y, Shi Y, Pirooznia M, Seifuddin F, et al. In vivo functional analysis of non-conserved human lncRNAs associated with cardiometabolic traits. Nat Commun. 2020;11(1):45.
Ruan X, et al. Identification of human long noncoding RNAs associated with nonalcoholic fatty liver disease and metabolic homeostasis. J Clin Invest. 2021;131(1):e136336.
Hennessy EJ, van Solingen C, Scacalossi KR, Ouimet M, Afonso MS, Prins J, et al. The long noncoding RNA CHROME regulates cholesterol homeostasis in primate. Nat Metab. 2019;1(1):98–110.
Zhang X, et al. Interrogation of nonconserved human adipose lincRNAs identifies a regulatory role of linc-ADAL in adipocyte metabolism. Sci Transl Med. 2018;10(446):eaar5987.
Tran KV, Brown EL, DeSouza T, Jespersen NZ, Nandrup-Bus C, Yang Q, et al. Human thermogenic adipocyte regulation by the long noncoding RNA LINC00473. Nat Metab. 2020;2(5):397–412.
Jin L, Lin X, Yang L, Fan X, Wang W, Li S, et al. AK098656, a novel vascular smooth muscle cell-dominant long noncoding RNA, promotes hypertension. Hypertension. 2018;71(2):262–72.
Seifuddin F, et al. lncRNAKB, a knowledgebase of tissue-specific functional annotation and trait association of long noncoding RNA. Sci Data. 2020;7(1):326.
Zhu Z, Zhang F, Hu H, Bakshi A, Robinson MR, Powell JE, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nat Genet. 2016;48(5):481–7.
Hasegawa M, Kawai K, Mitsui T, Taniguchi K, Monnai M, Wakui M, et al. The reconstituted 'humanized liver' in TK-NOG mice is mature and functional. Biochem Biophys Res Commun. 2011;405(3):405–10.
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–8.
Richardson TG, Sanderson E, Palmer TM, Ala-Korpela M, Ference BA, Davey Smith G, et al. Evaluating the relationship between circulating lipoprotein lipids and apolipoproteins with risk of coronary heart disease: a multivariable Mendelian randomisation analysis. PLoS Med. 2020;17(3):e1003062.
Frayn KN, Karpe F, Fielding BA, Macdonald IA, Coppack SW. Integrative physiology of human adipose tissue. Int J Obes Relat Metab Disord. 2003;27(8):875–88.
Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, et al. GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res. 2019;47(D1):D766–73.
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, et al. Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev. 2011;25(18):1915–27.
Mi L, Zhao XY, Li S, Yang G, Lin JD. Conserved function of the long noncoding RNA Blnc1 in brown adipocyte differentiation. Mol Metab. 2017;6(1):101–10.
Bell JL, Wächter K, Mühleck B, Pazaitis N, Köhn M, Lederer M, et al. Insulin-like growth factor 2 mRNA-binding proteins (IGF2BPs): post-transcriptional drivers of cancer progression? Cell Mol Life Sci. 2013;70(15):2657–75.
Diederichs S. The four dimensions of noncoding RNA conservation. Trends Genet. 2014;30(4):121–3.
Zhao XY, Li S, Wang GX, Yu Q, Lin JD. A long noncoding RNA transcriptional regulatory circuit drives thermogenic adipocyte differentiation. Mol Cell. 2014;55(3):372–82.
Zhao XY, Xiong X, Liu T, Mi L, Peng X, Rui C, et al. Long noncoding RNA licensing of obesity-linked hepatic lipogenesis and NAFLD pathogenesis. Nat Commun. 2018;9(1):2986.
Li S, Mi L, Yu L, Yu Q, Liu T, Wang GX, et al. Zbtb7b engages the long noncoding RNA Blnc1 to drive brown and beige fat development and thermogenesis. Proc Natl Acad Sci U S A. 2017;114(34):E7111–20.
Zhao XY, Li S, DelProposto JL, Liu T, Mi L, Porsche C, et al. The long noncoding RNA Blnc1 orchestrates homeostatic adipose tissue remodeling to preserve metabolic health. Mol Metab. 2018;14:60–70.
Yang L, Li P, Yang W, Ruan X, Kiesewetter K, Zhu J, et al. Integrative transcriptome analyses of metabolic responses in mice define pivotal LncRNA metabolic regulators. Cell Metab. 2016;24(4):627–39.
Liu C, Yang Z, Wu J, Zhang L, Lee S, Shin DJ, et al. Long noncoding RNA H19 interacts with polypyrimidine tract-binding protein 1 to reprogram hepatic lipid homeostasis. Hepatology. 2018;67(5):1768–83.
Schmidt E, Dhaouadi I, Gaziano I, Oliverio M, Klemm P, Awazawa M, et al. LincRNA H19 protects from dietary obesity by constraining expression of monoallelic genes in brown fat. Nat Commun. 2018;9(1):3622.
Tao SC, Rui BY, Wang QY, Zhou D, Zhang Y, Guo SC. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018;25(1):241–55.
Li DY, Busch A, Jin H, Chernogubova E, Pelisek J, Karlsson J, et al. H19 induces abdominal aortic aneurysm development and progression. Circulation. 2018;138(15):1551–68.
Sallam T, Jones MC, Gilliland T, Zhang L, Wu X, Eskin A, et al. Feedback modulation of cholesterol metabolism by the lipid-responsive non-coding RNA LeXis. Nature. 2016;534(7605):124–8.
Sallam T, Jones M, Thomas BJ, Wu X, Gilliland T, Qian K, et al. Transcriptional regulation of macrophage cholesterol efflux and atherogenesis by a long noncoding RNA. Nat Med. 2018;24(3):304–12.
Ding C, Lim YC, Chia SY, Walet ACE, Xu S, Lo KA, et al. De novo reconstruction of human adipose transcriptome reveals conserved lncRNAs as regulators of brown adipogenesis. Nat Commun. 2018;9(1):1329.
Kambarami RA, Chirenje M, Rusakaniko S, Anabwani G. Perinatal mortality rates and associated socio-demographic factors in two rural districts in Zimbabwe. Cent Afr J Med. 1997;43(6):158–62.
Ruan X, et al. Identification of human long non-coding RNAs associated with nonalcoholic fatty liver disease and metabolic homeostasis. J Clin Invest. 2020.
Matsui M, Corey DR. Non-coding RNAs as drug targets. Nat Rev Drug Discov. 2017;16(3):167–79.
Ransohoff JD, Wei Y, Khavari PA. The functions and unique features of long intergenic non-coding RNA. Nat Rev Mol Cell Biol. 2018;19(3):143–57.
Esguerra JL, Eliasson L. Functional implications of long non-coding RNAs in the pancreatic islets of Langerhans. Front Genet. 2014;5:209.
Ito R, Takahashi T, Ito M. Humanized mouse models: application to human diseases. J Cell Physiol. 2018;233(5):3723–8.
Walsh NC, Kenney LL, Jangalwe S, Aryee KE, Greiner DL, Brehm MA, et al. Humanized mouse models of clinical disease. Annu Rev Pathol. 2017;12:187–215.
Kim J, Koo BK, Knoblich JA. Human organoids: model systems for human biology and medicine. Nat Rev Mol Cell Biol. 2020;21(10):571–84.
Kadumuri RV, Janga SC. Epitranscriptomic code and its alterations in human disease. Trends Mol Med. 2018;24(10):886–903.
Zhang H, Shi X, Huang T, Zhao X, Chen W, Gu N, et al. Dynamic landscape and evolution of m6A methylation in human. Nucleic Acids Res. 2020;48(11):6251–64.
Liu J, et al. Landscape and regulation of m(6)A and m(6)Am methylome across human and mouse tissues. Mol Cell. 2020;77(2):426–40 e6.
Liu N, Zhou KI, Parisien M, Dai Q, Diatchenko L, Pan T. N6-methyladenosine alters RNA structure to regulate binding of a low-complexity protein. Nucleic Acids Res. 2017;45(10):6051–63.
Wu Y, et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol Cancer. 2019;18(1):87.
Ni W, Yao S, Zhou Y, Liu Y, Huang P, Zhou A, et al. Long noncoding RNA GAS5 inhibits progression of colorectal cancer by interacting with and triggering YAP phosphorylation and degradation and is negatively regulated by the m(6)A reader YTHDF3. Mol Cancer. 2019;18(1):143.
Zampetaki A, Albrecht A, Steinhofel K. Long non-coding RNA structure and function: is there a link? Front Physiol. 2018;9:1201.
Kwok CK, Tang Y, Assmann SM, Bevilacqua PC. The RNA structurome: transcriptome-wide structure probing with next-generation sequencing. Trends Biochem Sci. 2015;40(4):221–32.
Donlic A, Hargrove AE. Targeting RNA in mammalian systems with small molecules. Wiley Interdiscip Rev RNA. 2018;9(4):e1477.
Kornienko AE, Dotter CP, Guenzl PM, Gisslinger H, Gisslinger B, Cleary C, et al. Long non-coding RNAs display higher natural expression variation than protein-coding genes in healthy humans. Genome Biol. 2016;17:14.
Bhartiya D, Scaria V. Genomic variations in non-coding RNAs: structure, function and regulation. Genomics. 2016;107(2-3):59–68.
Funding
This study was funded by NHLBI Division of Intramural Research funds to HC (1ZIAHL006103, 1ZIAHL006159).
Author information
Authors and Affiliations
Contributions
Xiangbo Ruan initiated the idea for the article, performed the literature search, and drafted the manuscript.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The author declares no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article belongs to the Topical Collection: Translating genome medicine to treatments
Rights and permissions
About this article
Cite this article
Ruan, X. Targeting Human lncRNAs for Treating Cardiometabolic Diseases. Cardiovasc Drugs Ther 35, 655–662 (2021). https://doi.org/10.1007/s10557-021-07158-2
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s10557-021-07158-2